|
|
IV.A.4. (XIV.F.) |
Relative enthalpies of isomers - Comparison of 0K enthalpies (kJ mol-1)
Isomers of C2H2O
2015 06 30 15:39| index | Species | CAS number | Name | Relative experimental enthalpy (kJ mol-1) | sketch |
|---|---|---|---|---|---|
| a | C2H2O | 157186 | Oxirene | 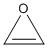 |
|
| b | CH2CO | 463514 | Ketene | 0.0 | 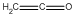 |
| c | HCCOH | 32038792 | ethynol | 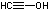 |
| composite | G2 | 314.7 a 0.0 b |
|---|---|---|
| G3 | 318.7 a 0.0 b |
|
| G4 | 0.0 b 141.3 c |
|
| CBS-Q | 316.6 a 0.0 b |
| STO-3G | 3-21G | 3-21G* | 6-31G | 6-31G* | 6-31G** | 6-31+G** | 6-311G* | 6-311G** | 6-31G(2df,p) | 6-311+G(3df,2p) | TZVP | cc-pVDZ | cc-pVTZ | cc-pVQZ | aug-cc-pVDZ | aug-cc-pVTZ | aug-cc-pVQZ | cc-pV(T+d)Z | daug-cc-pVDZ | daug-cc-pVTZ | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hartree fock | HF | 266.9 a 0.0 b 89.4 c |
382.5 a 0.0 b 132.2 c |
382.5 a 0.0 b 132.3 c |
358.4 a 0.0 b 132.2 c |
365.7 a 0.0 b 167.1 c |
365.0 a 0.0 b 154.5 c |
358.9 a 0.0 b 149.9 c |
378.8 a 0.0 b 168.2 c |
374.8 a 0.0 b 152.1 c |
370.4 a 0.0 b 156.2 c |
363.9 a 0.0 b 152.5 c |
363.9 a 0.0 b 147.7 c |
379.1 a 0.0 b 159.5 c |
366.4 a 0.0 b 151.3 c |
365.1 a 0.0 b 151.1 c |
363.7 a 0.0 b 154.1 c |
361.5 a 0.0 b 150.5 c |
362.8 a 0.0 b |
0.0 b 151.3 c |
364.2 a 0.0 b |
361.4 a 0.0 b |
| density functional | LSDA | 0.0 b 166.4 c |
0.0 b 163.8 c |
0.0 b 163.8 c |
0.0 b 162.6 c |
0.0 b 172.8 c |
0.0 b 162.3 c |
0.0 b 154.9 c |
0.0 b 171.1 c |
0.0 b 155.9 c |
0.0 b 157.7 c |
0.0 b 161.6 c |
0.0 b 151.2 c |
0.0 b 152.2 c |
0.0 b 151.2 c |
|||||||
| BLYP | 286.6 a 0.0 b 166.6 c |
0.0 b 171.3 c |
0.0 b 171.3 c |
0.0 b 169.6 c |
0.0 b 181.2 c |
0.0 b 170.9 c |
0.0 b 162.9 c |
0.0 b 177.7 c |
0.0 b 163.8 c |
0.0 b 166.4 c |
0.0 b 169.2 c |
0.0 b 159.8 c |
0.0 b 159.8 c |
|||||||||
| B1B95 | 276.6 a 0.0 b 145.1 c |
362.8 a 0.0 b 149.7 c |
362.8 a 0.0 b 149.7 c |
332.0 a 0.0 b 146.6 c |
331.8 a 0.0 b |
331.8 a 0.0 b 145.3 c |
322.7 a 0.0 b 148.0 c |
344.0 a 0.0 b 164.5 c |
340.6 a 0.0 b 149.4 c |
334.1 a 0.0 b 151.6 c |
326.4 a 0.0 b |
332.0 a 0.0 b |
0.0 b 155.8 c |
330.4 a 0.0 b 154.1 c |
324.3 a 0.0 b 155.4 c |
324.1 a 0.0 b |
0.0 b 154.1 c |
324.5 a 0.0 b |
324.0 a 0.0 b |
|||
| B3LYP | 282.3 a 0.0 b 150.8 c |
0.0 b 161.7 c |
0.0 b 161.7 c |
0.0 b 159.9 c |
0.0 b 176.0 c |
0.0 b 165.1 c |
317.3 a 0.0 b 158.0 c |
0.0 b 173.5 c |
0.0 b 158.9 c |
0.0 b 161.6 c |
0.0 b 156.3 c |
325.2 a 0.0 b 164.8 c |
0.0 b 155.5 c |
0.0 b 156.0 c |
0.0 b 153.6 c |
0.0 b 155.5 c |
||||||
| B3LYPultrafine | 0.0 b 175.9 c |
0.0 b 158.0 c |
0.0 b 155.5 c |
0.0 b 153.6 c |
||||||||||||||||||
| B3PW91 | 284.5 a 0.0 b 155.3 c |
367.9 a 0.0 b 161.9 c |
367.9 a 0.0 b 161.9 c |
0.0 b 160.6 c |
0.0 b 176.4 c |
0.0 b 165.2 c |
0.0 b 159.3 c |
0.0 b 175.8 c |
0.0 b 160.4 c |
0.0 b 162.0 c |
331.5 a 0.0 b |
0.0 b 166.3 c |
0.0 b 156.9 c |
327.6 a 0.0 b |
328.9 a 0.0 b |
0.0 b 156.8 c |
327.9 a 0.0 b |
328.8 a 0.0 b |
||||
| mPW1PW91 | 282.1 a 0.0 b 151.3 c |
368.4 a 0.0 b 159.7 c |
368.4 a 0.0 b 159.7 c |
336.3 a 0.0 b 158.6 c |
0.0 b 175.6 c |
335.5 a 0.0 b 164.1 c |
327.6 a 0.0 b 158.3 c |
0.0 b 175.3 c |
0.0 b 159.7 c |
339.0 a 0.0 b 161.2 c |
331.9 a 0.0 b |
336.5 a 0.0 b |
0.0 b 165.7 c |
335.6 a 0.0 b 156.2 c |
328.6 a 0.0 b |
329.4 a 0.0 b |
0.0 b 153.3 c |
328.9 a 0.0 b |
329.3 a 0.0 b |
|||
| M06-2X | 263.5 a 0.0 b |
350.3 a 0.0 b |
350.3 a 0.0 b 136.1 c |
324.8 a 0.0 b |
324.9 a 0.0 b 157.9 c |
323.8 a 0.0 b |
317.0 a 0.0 b |
337.9 a 0.0 b |
334.2 a 0.0 b |
328.8 a 0.0 b |
321.8 a 0.0 b |
323.9 a 0.0 b |
334.7 a 0.0 b |
324.4 a 0.0 b |
320.3 a 0.0 b |
319.1 a 0.0 b |
320.6 a 0.0 b |
319.1 a 0.0 b |
||||
| PBEPBE | 0.0 b 171.9 c |
0.0 b 172.1 c |
0.0 b 172.1 c |
0.0 b 171.1 c |
0.0 b 182.1 c |
0.0 b 171.2 c |
0.0 b 164.2 c |
0.0 b 180.6 c |
0.0 b 165.6 c |
0.0 b 167.1 c |
321.8 a 0.0 b |
0.0 b 171.1 c |
0.0 b 160.8 c |
0.0 b 158.6 c |
0.0 b 160.8 c |
|||||||
| PBEPBEultrafine | 0.0 b 182.1 c |
|||||||||||||||||||||
| PBE1PBE | 282.4 a 0.0 b |
368.1 a 0.0 b |
368.1 a 0.0 b |
336.5 a 0.0 b |
335.9 a 0.0 b 175.9 c |
335.9 a 0.0 b |
326.8 a 0.0 b |
344.2 a 0.0 b |
338.0 a 0.0 b |
330.4 a 0.0 b |
335.5 a 0.0 b |
334.4 a 0.0 b |
327.6 a 0.0 b |
327.9 a 0.0 b |
327.9 a 0.0 b |
327.8 a 0.0 b |
||||||
| HSEh1PBE | 283.0 a 0.0 b |
369.7 a 0.0 b 160.6 c |
369.7 a 0.0 b |
337.2 a 0.0 b |
0.0 b 175.9 c |
327.5 a 0.0 b 158.4 c |
339.2 a 0.0 b |
331.5 a 0.0 b |
336.4 a 0.0 b |
335.5 a 0.0 b 156.1 c |
328.4 a 0.0 b |
329.0 a 0.0 b |
328.7 a 0.0 b |
328.9 a 0.0 b |
||||||||
| TPSSh | 281.3 a 0.0 b |
325.2 a 0.0 b 183.1 c |
0.0 b 166.3 c |
0.0 b 169.1 c |
320.5 a 0.0 b |
332.5 a 0.0 b |
0.0 b 163.7 c |
317.9 a 0.0 b |
318.9 a 0.0 b |
317.8 a 0.0 b |
||||||||||||
| wB97X-D | 278.8 a 0.0 b |
370.2 a 0.0 b |
370.2 a 0.0 b 158.2 c |
336.6 a 0.0 b |
338.2 a 0.0 b 174.2 c |
337.3 a 0.0 b |
329.0 a 0.0 b 157.1 c |
347.2 a 0.0 b 158.3 c |
341.0 a 0.0 b |
334.1 a 0.0 b |
338.4 a 0.0 b 156.1 c |
0.0 b 167.9 c |
338.2 a 0.0 b 155.9 c |
336.1 a 0.0 b |
330.8 a 0.0 b |
332.2 a 0.0 b 154.4 c |
333.4 a 0.0 b |
331.0 a 0.0 b |
332.1 a 0.0 b |
|||
| B97D3 | 294.8 a 0.0 b |
0.0 b 170.7 c |
0.0 b 181.8 c |
0.0 b 164.4 c |
0.0 b 166.1 c |
0.0 b 161.8 c |
0.0 b 163.0 c |
0.0 b 162.0 c |
0.0 b 160.0 c |
326.3 a 0.0 b |
||||||||||||
| STO-3G | 3-21G | 3-21G* | 6-31G | 6-31G* | 6-31G** | 6-31+G** | 6-311G* | 6-311G** | 6-31G(2df,p) | 6-311+G(3df,2p) | TZVP | cc-pVDZ | cc-pVTZ | cc-pVQZ | aug-cc-pVDZ | aug-cc-pVTZ | aug-cc-pVQZ | cc-pV(T+d)Z | daug-cc-pVDZ | daug-cc-pVTZ | ||
| Moller Plesset perturbation | MP2 | 293.1 a 0.0 b 111.1 c |
370.9 a 0.0 b 146.1 c |
370.9 a 0.0 b 146.1 c |
330.5 a 0.0 b 140.3 c |
0.0 b 159.9 c |
0.0 b 152.3 c |
320.6 a 0.0 b |
0.0 b 165.1 c |
0.0 b 147.0 c |
335.9 a 0.0 b 147.1 c |
325.2 a 0.0 b |
0.0 b 145.7 c |
0.0 b 155.1 c |
330.4 a 0.0 b 141.4 c |
326.1 a 0.0 b |
0.0 b 146.8 c |
321.5 a 0.0 b |
0.0 b 141.4 c |
321.0 a 0.0 b |
||
| MP2=FULL | 293.1 a 0.0 b 111.2 c |
370.8 a 0.0 b 146.0 c |
370.8 a 0.0 b 146.0 c |
330.5 a 0.0 b 140.2 c |
0.0 b 160.2 c |
332.4 a 0.0 b 152.7 c |
321.9 a 0.0 b 147.5 c |
0.0 b 165.0 c |
0.0 b 146.8 c |
341.9 a 0.0 b 148.4 c |
327.3 a 0.0 b |
0.0 b 155.2 c |
332.5 a 0.0 b 141.6 c |
327.9 a 0.0 b |
325.2 a 0.0 b |
323.4 a 0.0 b |
324.9 a 0.0 b |
|||||
| MP3 | 320.7 a 0.0 b |
314.8 a 0.0 b |
324.4 a 0.0 b |
324.7 a 0.0 b |
345.5 a 0.0 b |
328.2 a 0.0 b |
326.1 a 0.0 b |
320.9 a 0.0 b |
||||||||||||||
| MP3=FULL | 344.8 a 0.0 b |
344.8 a 0.0 b |
310.3 a 0.0 b |
322.3 a 0.0 b 152.8 c |
323.9 a 0.0 b |
316.2 a 0.0 b 141.2 c |
341.9 a 0.0 b |
339.8 a 0.0 b |
338.5 a 0.0 b |
326.6 a 0.0 b |
325.2 a 0.0 b |
346.2 a 0.0 b |
330.4 a 0.0 b |
326.0 a 0.0 b |
325.5 a 0.0 b |
327.0 a 0.0 b |
325.2 a 0.0 b |
|||||
| MP4 | NC NC |
0.0 b 158.6 c |
0.0 b 169.5 c |
0.0 b 157.3 c |
||||||||||||||||||
| B2PLYP | 287.7 a 0.0 b |
0.0 b 171.0 c |
0.0 b 151.3 c |
|||||||||||||||||||
| B2PLYP=FULL | 287.7 a 0.0 b |
|||||||||||||||||||||
| B2PLYP=FULLultrafine | 287.7 a 0.0 b |
337.3 a 0.0 b |
350.7 a 0.0 b |
337.2 a 0.0 b |
329.6 a 0.0 b |
|||||||||||||||||
| Configuration interaction | CID | 359.3 a 0.0 b 133.0 c |
359.3 a 0.0 b |
326.5 a 0.0 b |
335.4 a 0.0 b 159.3 c |
354.2 a 0.0 b |
NC NC |
337.6 a 0.0 b |
358.0 a 0.0 b |
341.4 a 0.0 b |
339.2 a 0.0 b |
334.5 a 0.0 b |
||||||||||
| CISD | 361.2 a 0.0 b 137.2 c |
361.2 a 0.0 b |
328.1 a 0.0 b |
337.1 a 0.0 b 161.7 c |
355.7 a 0.0 b |
338.2 a 0.0 b |
359.3 a 0.0 b |
342.0 a 0.0 b |
339.4 a 0.0 b |
334.9 a 0.0 b |
||||||||||||
| STO-3G | 3-21G | 3-21G* | 6-31G | 6-31G* | 6-31G** | 6-31+G** | 6-311G* | 6-311G** | 6-31G(2df,p) | 6-311+G(3df,2p) | TZVP | cc-pVDZ | cc-pVTZ | cc-pVQZ | aug-cc-pVDZ | aug-cc-pVTZ | aug-cc-pVQZ | cc-pV(T+d)Z | daug-cc-pVDZ | daug-cc-pVTZ | ||
| Quadratic configuration interaction | QCISD | NC NC |
349.7 a 0.0 b 137.4 c |
349.7 a 0.0 b |
315.0 a 0.0 b 133.1 c |
326.3 a 0.0 b 159.5 c |
327.5 a 0.0 b 152.0 c |
318.2 a 0.0 b 146.6 c |
344.8 a 0.0 b 165.3 c |
342.8 a 0.0 b 148.3 c |
335.6 a 0.0 b 149.3 c |
326.1 a 0.0 b |
327.9 a 0.0 b |
0.0 b 156.9 c |
330.9 a 0.0 b 143.5 c |
325.3 a 0.0 b |
322.9 a 0.0 b |
326.1 a 0.0 b |
322.4 a 0.0 b |
|||
| QCISD(T) | 0.0 b 160.0 c |
319.6 a 0.0 b |
0.0 b 143.7 c |
316.0 a 0.0 b |
315.4 a 0.0 b |
|||||||||||||||||
| QCISD(T)=FULL | 313.5 a 0.0 b |
322.0 a 0.0 b |
322.9 a 0.0 b |
320.2 a 0.0 b |
318.6 a 0.0 b |
319.8 a 0.0 b |
||||||||||||||||
| Coupled Cluster | CCD | NC NC |
346.4 a 0.0 b 128.3 c |
346.4 a 0.0 b |
312.1 a 0.0 b 123.5 c |
322.1 a 0.0 b 152.9 c |
323.5 a 0.0 b 145.9 c |
315.7 a 0.0 b 141.2 c |
341.6 a 0.0 b 158.8 c |
339.6 a 0.0 b 142.2 c |
332.9 a 0.0 b 143.6 c |
324.5 a 0.0 b |
325.3 a 0.0 b |
346.0 a 0.0 b 150.3 c |
328.6 a 0.0 b 138.3 c |
324.9 a 0.0 b |
321.7 a 0.0 b |
325.6 a 0.0 b |
||||
| CCSD | 323.8 a 0.0 b 155.9 c |
325.0 a 0.0 b |
316.3 a 0.0 b |
343.1 a 0.0 b |
340.9 a 0.0 b 145.3 c |
333.8 a 0.0 b |
324.8 a 0.0 b |
326.0 a 0.0 b |
347.0 a 0.0 b |
329.3 a 0.0 b 140.9 c |
326.5 a 0.0 b |
324.3 a 0.0 b |
321.8 a 0.0 b |
325.1 a 0.0 b |
||||||||
| CCSD=FULL | 325.5 a 0.0 b |
340.1 a 0.0 b |
327.3 a 0.0 b |
326.7 a 0.0 b |
347.7 a 0.0 b |
331.7 a 0.0 b |
325.1 a 0.0 b |
326.0 a 0.0 b |
326.1 a 0.0 b |
325.7 a 0.0 b |
||||||||||||
| CCSD(T) | 319.4 a 0.0 b 159.0 c |
320.8 a 0.0 b |
312.0 a 0.0 b |
0.0 b 147.9 c |
328.3 a 0.0 b |
319.3 a 0.0 b |
324.0 a 0.0 b 142.7 c |
320.7 a 0.0 b |
318.4 a 0.0 b |
315.8 a 0.0 b |
316.4 a 0.0 b |
319.4 a 0.0 b |
315.2 a 0.0 b |
|||||||||
| CCSD(T)=FULL | 321.2 a 0.0 b |
321.7 a 0.0 b |
326.3 a 0.0 b |
322.6 a 0.0 b |
319.3 a 0.0 b |
320.0 a 0.0 b |
318.4 a 0.0 b |
320.4 a 0.0 b |
319.6 a 0.0 b |
|||||||||||||
| STO-3G | 3-21G | 3-21G* | 6-31G | 6-31G* | 6-31G** | 6-31+G** | 6-311G* | 6-311G** | 6-31G(2df,p) | 6-311+G(3df,2p) | TZVP | cc-pVDZ | cc-pVTZ | cc-pVQZ | aug-cc-pVDZ | aug-cc-pVTZ | aug-cc-pVQZ | cc-pV(T+d)Z | daug-cc-pVDZ | daug-cc-pVTZ |
| CEP-31G | CEP-31G* | CEP-121G | CEP-121G* | LANL2DZ | SDD | cc-pVTZ-PP | aug-cc-pVTZ-PP | Def2TZVPP | ||
|---|---|---|---|---|---|---|---|---|---|---|
| hartree fock | HF | 336.7 a 0.0 b 139.9 c |
346.9 a 0.0 b 181.7 c |
338.3 a 0.0 b 118.0 c |
351.8 a 0.0 b 157.4 c |
354.5 a 0.0 b 119.4 c |
354.7 a 0.0 b 119.0 c |
365.2 a 0.0 b 151.0 c |
||
| density functional | B1B95 | 0.0 b 157.5 c |
0.0 b 178.5 c |
328.8 a 0.0 b |
||||||
| B3LYP | 309.1 a 0.0 b 162.0 c |
0.0 b 183.4 c |
311.7 a 0.0 b 147.3 c |
0.0 b 166.2 c |
0.0 b 147.8 c |
0.0 b 148.3 c |
0.0 b 155.4 c |
|||
| B3PW91 | 333.3 a 0.0 b |
|||||||||
| mPW1PW91 | 334.0 a 0.0 b |
|||||||||
| M06-2X | 322.5 a 0.0 b |
|||||||||
| PBEPBE | 0.0 b 160.8 c |
|||||||||
| PBE1PBE | 332.6 a 0.0 b |
|||||||||
| HSEh1PBE | 333.8 a 0.0 b |
|||||||||
| wB97X-D | 310.9 a 0.0 b |
318.9 a 0.0 b |
314.2 a 0.0 b |
324.0 a 0.0 b |
331.4 a 0.0 b |
331.9 a 0.0 b |
336.7 a 0.0 b |
|||
| Moller Plesset perturbation | MP2 | 309.4 a 0.0 b 145.3 c |
314.3 a 0.0 b 172.4 c |
310.1 a 0.0 b 131.2 c |
314.6 a 0.0 b 156.4 c |
327.4 a 0.0 b 129.4 c |
327.3 a 0.0 b 129.5 c |
328.1 a 0.0 b 141.8 c |
||
| MP2=FULL | 329.1 a 0.0 b |
|||||||||
| MP3 | 326.5 a 0.0 b |
|||||||||
| MP3=FULL | 327.5 a 0.0 b |
|||||||||
| Configuration interaction | CID | 339.7 a 0.0 b |
||||||||
| CISD | 340.3 a 0.0 b |
|||||||||
| Quadratic configuration interaction | QCISD | 328.8 a 0.0 b |
||||||||
| QCISD(T) | 322.1 a 0.0 b |
|||||||||
| QCISD(T)=FULL | 323.3 a 0.0 b |
|||||||||
| Coupled Cluster | CCD | 326.8 a 0.0 b |
||||||||
| CCSD | 327.3 a 0.0 b |
|||||||||
| CCSD=FULL | 328.4 a 0.0 b |
|||||||||
| CCSD(T) | 321.8 a 0.0 b |
|||||||||
| CCSD(T)=FULL | 322.9 a 0.0 b |
For descriptions of the methods (AM1, HF, MP2, ...) and basis sets (3-21G, 3-21G*, 6-31G, ...) see the glossary in section I.C. Predefined means the basis set used is determined by the method.
gaw refers to the group additivity method implemeted in the NIST Chemistry Webbook.
See section III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.