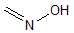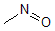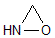Methods with standard basis sets
|
|
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
cc-pVQZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
aug-cc-pVQZ |
daug-cc-pVTZ |
| hartree fock |
HF |
0.0 a
102.4 b
98.3 c
123.8 d
34.5 e |
0.0 a
223.5 b
308.0 c
347.2 d
72.3 e |
0.0 a
223.5 b
308.0 c
347.2 d
72.3 e |
0.0 a
237.0 b
293.7 c
380.1 d
75.8 e |
0.0 a
234.5 b
261.3 c
318.6 d
77.9 e |
0.0 a
234.2 b
273.3 c
325.4 d
56.3 e |
0.0 a
235.4 b
278.4 c
333.1 d
56.2 e |
0.0 a
239.6 b
266.4 c
326.7 d
67.7 e |
0.0 a
234.0 b
275.5 c
330.5 d
56.1 e |
0.0 a
234.9 b
270.4 c
322.5 d
55.6 e |
0.0 a
78.3 e |
0.0 a
234.1 b
277.2 c
332.2 d
56.7 e |
0.0 a
229.5 b
266.1 c
324.7 d
54.4 e |
0.0 a
235.2 b
276.6 c
328.1 d
54.7 e |
0.0 a
236.6 b
278.2 c
330.3 d
54.6 e |
0.0 a
231.9 b
271.5 c
327.8 d
52.3 e |
0.0 a
236.2 b
277.9 c
328.9 d
54.0 e |
0.0 a
236.8 b
278.4 c
330.5 d
54.4 e |
0.0 a
236.1 b
277.7 c
328.8 d
53.9 e |
| density functional |
LSDA |
NC
NC
NC |
0.0 a
231.8 b
302.6 c
329.3 d
81.3 e |
NC
NC
NC |
NC
NC
NC |
0.0 a
215.5 b
259.5 c
285.0 d
60.5 e |
0.0 a
215.4 b
269.0 c
290.3 d
54.8 e |
0.0 a
216.4 b
275.1 c
299.3 d
55.6 e |
0.0 a
227.4 b
272.1 c
304.4 d
67.3 e |
0.0 a
221.6 b
280.7 c
307.8 d
56.3 e |
0.0 a
215.7 b
270.0 c
287.1 d
54.0 e |
|
NC
NC
NC |
0.0 a
213.1 b
266.1 c
293.5 d
54.1 e |
0.0 a
218.1 b
280.4 c
299.7 d
53.4 e |
|
0.0 a
213.9 b
274.7 c
295.2 d
51.6 e |
0.0 a
217.9 b
281.2 c
299.0 d
52.2 e |
|
|
| BLYP |
0.0 a
89.6 b
79.7 c
117.8 d
31.0 e |
0.0 a
199.4 b
258.0 c
300.4 d
76.5 e |
0.0 a
199.4 b
258.0 c
300.4 d
76.5 e |
0.0 a
207.2 b
254.4 c
323.4 d
80.1 e |
0.0 a
197.8 b
230.0 c
280.2 d
59.6 e |
0.0 a
198.7 b
240.4 c
286.4 d
55.2 e |
0.0 a
200.0 b
248.9 c
296.9 d
55.8 e |
0.0 a
208.2 b
242.6 c
299.4 d
66.2 e |
0.0 a
203.0 b
250.4 c
302.5 d
56.4 e |
0.0 a
200.5 b
243.2 c
286.0 d
54.5 e |
|
0.0 a
204.6 b
256.2 c
306.4 d
57.6 e |
0.0 a
196.7 b
238.2 c
290.5 d
54.8 e |
0.0 a
202.6 b
254.0 c
298.8 d
54.6 e |
|
0.0 a
198.2 b
249.8 c
293.9 d
71.7 e |
0.0 a
202.4 b
256.0 c
298.8 d
72.5 e |
|
|
| B1B95 |
0.0 a
104.8 b
97.4 c
118.9 d
35.5 e |
0.0 a
218.4 b
285.7 c
314.1 d
78.1 e |
0.0 a
218.4 b
285.7 c
314.1 d
78.1 e |
0.0 a
227.0 b
277.6 c
339.1 d
81.3 e |
0.0 a
214.7 b
247.9 c
280.1 d
60.2 e |
0.0 a
214.7 b
247.9 c
280.1 d
60.2 e |
0.0 a
215.9 b
264.6 c
294.7 d
54.7 e |
0.0 a
223.7 b
257.1 c
294.2 d
65.9 e |
0.0 a
218.2 b
265.8 c
297.8 d
54.9 e |
0.0 a
215.1 b
258.5 c
283.0 d
53.1 e |
|
0.0 a
219.3 b
269.3 c
301.5 d
56.3 e |
0.0 a
212.4 b
254.8 c
288.3 d
53.5 e |
0.0 a
216.6 b
266.7 c
291.6 d
52.8 e |
|
0.0 a
213.5 b
262.9 c
290.6 d
50.8 e |
0.0 a
217.1 b
268.4 c
291.9 d
52.0 e |
|
|
| B3LYP |
0.0 a
97.5 b
90.3 c
121.1 d
33.6 e |
0.0 a
210.8 b
276.6 c
315.9 d
77.6 e |
0.0 a
210.8 b
276.6 c
315.9 d
77.6 e |
0.0 a
219.4 b
269.9 c
340.0 d
80.7 e |
0.0 a
209.8 b
243.4 c
290.3 d
61.0 e |
0.0 a
209.6 b
253.3 c
295.8 d
55.2 e |
0.0 a
211.2 b
261.2 c
305.9 d
55.8 e |
0.0 a
218.8 b
254.2 c
306.7 d
66.6 e |
0.0 a
213.4 b
262.5 c
310.2 d
56.2 e |
0.0 a
210.9 b
254.9 c
294.8 d
54.4 e |
0.0 a
75.0 e |
0.0 a
215.0 b
267.3 c
314.0 d
57.4 e |
0.0 a
207.1 b
250.3 c
298.8 d
54.9 e |
0.0 a
213.2 b
265.3 c
306.5 d
54.4 e |
0.0 a
214.5 b
267.4 c
308.3 d
54.0 e |
0.0 a
209.0 b
260.8 c
302.7 d
52.1 e |
0.0 a
213.5 b
267.2 c
306.8 d
53.3 e |
0.0 a
214.2 b
267.6 c
308.3 d
53.5 e |
|
| B3LYPultrafine |
|
NC
NC
NC
NC |
|
|
0.0 a
209.8 b
243.4 c
290.3 d
61.0 e |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
|
|
|
NC
NC
NC
NC |
0.0 a
207.1 b
250.3 c
298.8 d
54.9 e |
0.0 a
213.3 b
265.3 c
306.5 d
54.4 e |
|
0.0 a
209.0 b
260.8 c
302.8 d
52.1 e |
0.0 a
213.6 b
267.2 c
306.8 d
53.3 e |
|
|
| B3PW91 |
0.0 a
103.4 b
93.8 c
121.6 d
36.1 e |
0.0 a
218.1 b
284.2 c
316.7 d
79.5 e |
0.0 a
218.1 b
284.2 c
316.7 d
79.5 e |
0.0 a
225.9 b
275.5 c
340.2 d
82.7 e |
0.0 a
213.8 b
246.8 c
285.0 d
61.9 e |
0.0 a
213.7 b
257.2 c
290.8 d
56.0 e |
0.0 a
214.8 b
263.4 c
299.0 d
56.4 e |
0.0 a
223.3 b
257.0 c
299.5 d
67.7 e |
0.0 a
217.7 b
265.6 c
303.1 d
56.6 e |
0.0 a
214.1 b
257.5 c
287.9 d
54.7 e |
|
0.0 a
218.4 b
268.6 c
305.9 d
57.8 e |
0.0 a
211.2 b
253.2 c
292.3 d
55.2 e |
0.0 a
216.1 b
266.5 c
297.4 d
54.4 e |
|
0.0 a
212.3 b
261.3 c
294.3 d
73.5 e |
0.0 a
216.4 b
267.9 c
297.4 d
74.2 e |
|
|
| mPW1PW91 |
0.0 a
104.4 b
95.5 c
120.5 d
36.0 e |
0.0 a
220.0 b
287.8 c
318.0 d
79.2 e |
0.0 a
220.0 b
287.8 c
318.0 d
79.2 e |
0.0 a
228.0 b
278.1 c
342.0 d
82.5 e |
0.0 a
215.8 b
248.7 c
285.0 d
61.8 e |
0.0 a
215.7 b
259.3 c
291.1 d
55.8 e |
0.0 a
217.0 b
265.5 c
299.6 d
56.4 e |
0.0 a
225.2 b
258.4 c
299.1 d
67.7 e |
0.0 a
219.6 b
267.2 c
302.8 d
56.5 e |
0.0 a
216.0 b
259.3 c
287.8 d
54.4 e |
|
0.0 a
220.2 b
270.1 c
305.8 d
57.7 e |
0.0 a
213.3 b
255.2 c
292.6 d
55.1 e |
0.0 a
218.0 b
268.0 c
297.2 d
54.3 e |
|
0.0 a
214.3 b
263.0 c
294.6 d
52.5 e |
0.0 a
218.4 b
269.6 c
297.4 d
53.4 e |
|
|
| M06-2X |
0.0 a
102.4 b
97.5 c
115.2 d
33.4 e |
0.0 a
216.0 b
290.4 c
313.2 d
72.0 e |
0.0 a
216.0 b
290.4 c
313.2 d
72.0 e |
0.0 a
223.8 b
280.8 c
341.7 d
75.1 e |
0.0 a
209.7 b
246.6 c
277.5 d
53.6 e |
0.0 a
209.1 b
256.4 c
283.1 d
47.5 e |
0.0 a
210.6 b
262.7 c
292.1 d
48.4 e |
0.0 a
219.7 b
257.5 c
291.4 d
59.8 e |
0.0 a
213.9 b
265.9 c
294.9 d
48.6 e |
0.0 a
210.9 b
257.8 c
281.0 d
46.9 e |
0.0 a
269.5 c |
0.0 a
214.6 b
269.1 c
298.1 d
49.5 e |
0.0 a
206.5 b
254.2 c
285.0 d
45.9 e |
0.0 a
213.8 b
268.1 c
290.4 d
46.1 e |
|
0.0 a
208.8 b
262.4 c
288.9 d
44.4 e |
0.0 a
214.5 b
269.4 c
291.1 d
45.4 e |
|
|
| PBEPBE |
0.0 a
99.0 b
88.5 c
119.9 d
33.7 e |
0.0 a
210.1 b
269.0 c
302.1 d
78.7 e |
0.0 a
210.1 b
269.0 c
302.1 d
78.7 e |
0.0 a
217.8 b
263.0 c
326.1 d
82.8 e |
0.0 a
205.4 b
235.8 c
275.3 d
61.7 e |
0.0 a
205.1 b
245.7 c
280.7 d
55.8 e |
0.0 a
206.3 b
252.6 c
290.2 d
56.6 e |
0.0 a
215.1 b
246.4 c
291.5 d
67.4 e |
0.0 a
209.7 b
254.9 c
294.9 d
56.8 e |
0.0 a
205.5 b
246.8 c
277.6 d
54.5 e |
0.0 a
75.1 e |
0.0 a
210.9 b
259.7 c
298.7 d
58.2 e |
0.0 a
203.4 b
243.0 c
283.9 d
55.4 e |
0.0 a
207.7 b
256.8 c
288.7 d
54.5 e |
|
0.0 a
203.8 b
251.7 c
285.3 d
52.7 e |
0.0 a
207.7 b
258.6 c
288.7 d
53.4 e |
|
|
| PBEPBEultrafine |
|
NC
NC
NC
NC |
|
|
0.0 a
205.4 b
235.8 c
275.3 d
61.6 e |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
|
|
|
NC
NC
NC
NC |
0.0 a
203.4 b
243.0 c
284.0 d
55.4 e |
0.0 a
207.7 b
256.8 c
288.7 d
54.5 e |
|
0.0 a
203.7 b
251.7 c
285.3 d
52.7 e |
0.0 a
207.8 b
258.6 c
288.7 d
53.4 e |
|
|
| PBE1PBE |
0.0 a
105.0 b
96.1 c
119.8 d
35.9 e |
0.0 a
220.4 b
287.5 c
316.7 d
79.2 e |
0.0 a
220.4 b
287.5 c
316.7 d
79.2 e |
0.0 a
228.9 b
278.2 c
342.0 d
82.7 e |
0.0 a
215.9 b
248.0 c
283.4 d
61.8 e |
0.0 a
215.9 b
248.0 c
283.4 d
61.8 e |
0.0 a
217.0 b
264.9 c
298.1 d
56.3 e |
0.0 a
225.1 b
257.4 c
297.3 d
67.6 e |
0.0 a
219.5 b
266.4 c
301.1 d
56.3 e |
0.0 a
215.9 b
258.4 c
285.7 d
54.2 e |
|
0.0 a
220.3 b
269.9 c
304.5 d
57.6 e |
0.0 a
213.5 b
254.9 c
291.4 d
55.0 e |
0.0 a
217.8 b
267.5 c
295.2 d
54.0 e |
|
0.0 a
214.2 b
262.3 c
293.0 d
52.2 e |
0.0 a
218.2 b
269.2 c
295.5 d
53.1 e |
|
|
| HSEh1PBE |
0.0 a
104.4 b
95.5 c
120.3 d
35.5 e |
0.0 a
220.8 b
287.8 c
319.0 d
79.5 e |
0.0 a
220.8 b
287.8 c
319.0 d
79.5 e |
0.0 a
228.5 b
278.0 c
343.0 d
82.6 e |
0.0 a
215.7 b
247.8 c
284.9 d
61.8 e |
0.0 a
215.7 b
258.5 c
291.0 d
55.8 e |
NC
NC
NC |
0.0 a
224.8 b
257.4 c
299.2 d
67.5 e |
0.0 a
219.3 b
266.3 c
302.9 d
56.4 e |
0.0 a
216.0 b
258.5 c
287.8 d
54.6 e |
|
0.0 a
220.4 b
270.0 c
306.5 d
57.8 e |
0.0 a
213.2 b
254.5 c
292.9 d
55.2 e |
0.0 a
218.0 b
267.5 c
297.4 d
54.4 e |
|
0.0 a
214.0 b
262.3 c
294.7 d
52.4 e |
0.0 a
218.4 b
269.2 c
297.6 d
53.5 e |
|
|
| TPSSh |
|
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
0.0 a
203.7 b
231.9 c
265.1 d
74.2 e |
NC
NC
NC
NC |
0.0 a
205.0 b
248.4 c
279.1 d
71.3 e |
NC
NC
NC
NC |
|
0.0 a
54.7 e |
|
NC
NC
NC
NC |
NC
NC
NC
NC |
0.0 a
206.4 b
251.2 c
276.9 d
66.8 e |
|
NC
NC
NC
NC |
NC
NC
NC
NC |
|
|
| wB97X-D |
NC
NC
NC
NC |
NC
NC
NC
NC |
0.0 a
223.1 b
288.0 c
325.6 d
82.4 e |
NC
NC
NC
NC |
0.0 a
216.7 b
248.8 c
288.6 d
62.7 e |
NC
NC
NC
NC |
0.0 a
217.8 b
265.4 c
303.0 d
56.9 e |
NC
NC
NC
NC |
0.0 a
220.2 b
266.9 c
306.3 d
57.5 e |
NC
NC
NC
NC |
|
0.0 a
221.3 b
269.1 c
309.0 d
58.8 e |
0.0 a
223.0 b
263.9 c
304.2 d
56.9 e |
0.0 a
219.3 b
267.1 c
300.7 d
55.4 e |
NC
NC
NC
NC |
NC
NC
NC
NC |
0.0 a
219.8 b
268.4 c
301.1 d
54.6 e |
NC
NC
NC
NC |
|
| B97D3 |
|
0.0 a
77.7 e |
|
|
|
|
0.0 a
56.0 e |
|
0.0 a
56.3 e |
|
0.0 a
211.9 b
300.7 d
54.8 e |
0.0 a
57.4 e |
|
0.0 a
54.5 e |
|
|
0.0 a
53.5 e |
|
0.0 a
258.5 c |
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
cc-pVQZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
aug-cc-pVQZ |
daug-cc-pVTZ |
|---|
| Moller Plesset perturbation |
MP2 |
0.0 a
103.6 b
42.7 c
140.7 d
40.3 e |
0.0 a
209.4 b
252.0 c
314.7 d
74.8 e |
0.0 a
209.4 b
252.0 c
314.7 d
74.8 e |
0.0 a
216.1 b
256.1 c
339.0 d
76.2 e |
0.0 a
217.9 b
248.9 c
291.1 d
58.6 e |
0.0 a
217.7 b
256.9 c
294.8 d
53.6 e |
0.0 a
217.9 b
265.6 c
303.6 d
54.1 e |
0.0 a
227.5 b
252.8 c
303.6 d
65.9 e |
0.0 a
218.5 b
260.6 c
303.8 d
49.4 e |
NC
NC
NC
NC |
|
0.0 a
217.5 b
264.2 c
306.3 d
51.2 e |
0.0 a
214.7 b
251.2 c
298.1 d
49.9 e |
0.0 a
220.9 b
271.5 c
297.9 d
49.9 e |
0.0 a
223.4 b
276.7 c
301.3 d
50.1 e |
0.0 a
217.5 b
265.8 c
299.6 d
49.3 e |
0.0 a
221.5 b
274.2 c
298.3 d
49.7 e |
0.0 a
223.0 b
277.2 c
301.1 d
49.8 e |
|
| MP2=FULL |
0.0 a
103.6 b
42.6 c
140.7 d
40.3 e |
0.0 a
209.5 b
252.2 c
314.9 d
74.8 e |
0.0 a
209.5 b
252.2 c
314.9 d
74.8 e |
0.0 a
216.1 b
256.2 c
339.0 d
76.2 e |
0.0 a
218.8 b
250.1 c
292.0 d
58.9 e |
NC
NC
NC
NC |
0.0 a
219.0 b
266.7 c
304.6 d
54.7 e |
0.0 a
228.5 b
254.0 c
304.5 d
66.3 e |
0.0 a
219.6 b
261.9 c
304.7 d
49.9 e |
0.0 a
227.1 b
266.3 c
295.9 d
54.7 e |
|
0.0 a
218.1 b
265.1 c
306.8 d
51.5 e |
0.0 a
215.3 b
251.9 c
298.7 d
50.3 e |
0.0 a
223.3 b
274.1 c
299.8 d
50.7 e |
0.0 a
225.3 b
279.1 c
303.5 d
50.9 e |
0.0 a
218.5 b
266.6 c
300.0 d
49.6 e |
0.0 a
224.0 b
275.8 c
299.9 d
50.7 e |
0.0 a
225.0 b
303.2 d
50.6 e |
|
| MP3 |
|
|
|
|
0.0 a
203.2 b
243.5 c
282.9 d
50.2 e |
|
NC
NC
NC
NC |
|
|
|
|
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
|
|
|
|
|
| MP3=FULL |
|
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
|
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
|
NC
NC
NC
NC |
NC
NC
NC |
|
|
| MP4 |
|
0.0 a
197.0 b
242.7 c
301.0 d
72.8 e |
|
|
0.0 a
206.5 b
231.7 c
284.2 d
57.0 e |
|
|
|
0.0 a
207.3 b
241.4 c
295.0 d
49.1 e |
|
|
0.0 a
205.3 b
244.5 c
296.1 d
50.8 e |
0.0 a
204.0 b
233.4 c
290.3 d
49.7 e |
0.0 a
210.0 b
252.4 c
290.1 d
48.9 e |
|
NC
NC
NC
NC |
NC
NC
NC
NC |
|
|
| MP4=FULL |
|
0.0 a
197.2 b
243.0 c
301.2 d
72.9 e |
|
|
0.0 a
207.7 b
233.2 c
285.4 d
57.5 e |
|
|
|
0.0 a
208.6 b
242.9 c
296.1 d
49.6 e |
|
|
|
0.0 a
204.7 b
234.2 c
291.0 d
50.0 e |
0.0 a
213.6 b
256.3 c
293.2 d
50.8 e |
|
0.0 a
207.5 b
248.4 c
291.1 d
49.9 e |
0.0 a
214.2 b
258.0 c
292.6 d
51.3 e |
|
|
| B2PLYP |
0.0 a
100.9 b
73.8 c
128.8 d
43.3 e |
0.0 a
212.0 b
269.4 c
316.8 d
86.6 e |
0.0 a
212.0 b
269.4 c
316.8 d
86.6 e |
0.0 a
220.2 b
265.7 c
341.5 d
93.1 e |
0.0 a
214.0 b
244.7 c
292.3 d
74.0 e |
0.0 a
214.1 b
254.6 c
297.7 d
68.3 e |
0.0 a
215.0 b
262.6 c
307.1 d
71.5 e |
0.0 a
223.7 b
253.6 c
307.8 d
82.1 e |
0.0 a
217.2 b
262.1 c
310.3 d
68.5 e |
0.0 a
216.1 b
256.9 c
296.0 d
66.5 e |
|
0.0 a
217.8 b
266.6 c
313.5 d
71.8 e |
0.0 a
210.7 b
249.8 c
300.1 d
64.5 e |
0.0 a
216.6 b
266.3 c
304.7 d
66.1 e |
|
0.0 a
212.7 b
261.5 c
302.9 d
65.2 e |
0.0 a
216.9 b
268.4 c
304.9 d
66.3 e |
|
|
| B2PLYP=FULL |
0.0 a
100.9 b
73.8 c
128.8 d
43.4 e |
0.0 a
212.1 b
269.5 c
316.9 d
86.6 e |
0.0 a
212.1 b
269.5 c
316.9 d
86.6 e |
0.0 a
220.3 b
265.8 c
341.6 d
93.2 e |
0.0 a
214.4 b
245.2 c
292.7 d
74.3 e |
0.0 a
214.4 b
254.9 c
298.0 d
68.4 e |
0.0 a
215.2 b
262.9 c
307.4 d
71.7 e |
0.0 a
223.9 b
253.9 c
308.0 d
82.1 e |
0.0 a
217.4 b
262.4 c
310.5 d
68.6 e |
0.0 a
217.2 b
257.6 c
296.6 d
67.0 e |
|
0.0 a
217.9 b
266.9 c
313.6 d
71.9 e |
0.0 a
210.9 b
250.1 c
300.3 d
64.6 e |
0.0 a
217.3 b
267.0 c
305.3 d
66.4 e |
|
0.0 a
213.0 b
261.7 c
303.0 d
65.3 e |
0.0 a
217.5 b
268.8 c
305.3 d
66.6 e |
|
|
| B2PLYP=FULLultrafine |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
|
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
|
NC
NC
NC |
|
|
|
NC
NC
NC |
|
|
|
| Configuration interaction |
CID |
|
0.0 a
198.4 b
268.5 c
312.5 d
66.6 e |
0.0 a
198.4 b
268.5 c
312.5 d
66.6 e |
0.0 a
209.8 b
265.6 c
343.1 d
68.9 e |
0.0 a
216.1 b
250.7 c
294.2 d
56.4 e |
|
|
0.0 a
224.2 b
253.2 c
303.9 d
63.1 e |
|
|
|
|
|
|
|
|
|
|
|
| CISD |
|
0.0 a
200.5 b
268.7 c
313.2 d
68.4 e |
0.0 a
200.5 b
268.7 c
313.2 d
68.4 e |
0.0 a
211.4 b
265.2 c
343.3 d
70.7 e |
0.0 a
216.8 b
249.8 c
294.8 d
57.3 e |
|
|
0.0 a
225.0 b
252.3 c
304.7 d
64.0 e |
|
|
|
|
|
|
|
|
|
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
cc-pVQZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
aug-cc-pVQZ |
daug-cc-pVTZ |
|---|
| Quadratic configuration interaction |
QCISD |
|
0.0 a
188.0 b
250.6 c
300.2 d
66.5 e |
0.0 a
188.0 b
250.6 c
300.2 d
66.5 e |
0.0 a
198.4 b
251.8 c
330.0 d
68.7 e |
0.0 a
204.6 b
237.8 c
286.6 d
54.0 e |
0.0 a
204.7 b
245.5 c
290.1 d
49.8 e |
0.0 a
205.3 b
252.8 c
298.5 d
50.4 e |
0.0 a
213.5 b
239.2 c
297.0 d
61.3 e |
0.0 a
205.7 b
246.3 c
297.3 d
46.9 e |
0.0 a
211.6 b
251.0 c
291.0 d
49.7 e |
|
0.0 a
204.0 b
249.0 c
298.7 d
48.3 e |
0.0 a
201.9 b
237.8 c
292.2 d
47.4 e |
0.0 a
210.5 b
257.8 c
295.5 d
47.3 e |
|
0.0 a
205.4 b
250.6 c
295.3 d
47.2 e |
NC
NC
NC
NC |
|
|
| QCISD(T) |
|
|
|
|
0.0 a
199.2 b
231.0 c
280.5 d
52.4 e |
|
|
NC
NC
NC
NC |
|
|
|
0.0 a
199.1 b
243.9 c
293.4 d
46.6 e |
0.0 a
197.3 b
232.1 c
287.0 d
45.6 e |
0.0 a
204.9 b
252.5 c
289.1 d
45.3 e |
|
NC
NC
NC
NC |
0.0 a
206.2 b
255.6 c
289.9 d
46.1 e |
|
|
| QCISD(T)=FULL |
|
|
|
|
NC
NC
NC
NC |
|
NC
NC
NC
NC |
|
|
|
|
|
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
|
|
| Coupled Cluster |
CCD |
|
0.0 a
183.4 b
250.7 c
298.0 d
62.2 e |
0.0 a
183.4 b
250.7 c
298.0 d
62.2 e |
0.0 a
194.6 b
252.1 c
328.7 d
64.1 e |
0.0 a
202.0 b
238.4 c
283.0 d
51.1 e |
0.0 a
202.0 b
245.7 c
286.6 d
46.7 e |
0.0 a
203.4 b
253.6 c
295.7 d
47.5 e |
0.0 a
210.7 b
240.0 c
293.2 d
58.3 e |
0.0 a
202.8 b
246.7 c
293.6 d
43.9 e |
0.0 a
209.6 b
251.9 c
288.6 d
47.2 e |
|
0.0 a
201.7 b
249.4 c
295.8 d
45.4 e |
0.0 a
198.7 b
237.6 c
288.4 d
44.2 e |
0.0 a
208.9 b
259.3 c
293.4 d
45.0 e |
|
0.0 a
203.7 b
251.4 c
293.8 d
44.4 e |
0.0 a
211.0 b
262.4 c
295.0 d
46.0 e |
|
|
| CCSD |
|
|
|
|
0.0 a
202.7 b
237.0 c
284.4 d
52.2 e |
|
|
|
|
0.0 a
210.0 b
250.3 c
289.4 d
71.0 e |
|
0.0 a
202.4 b
248.2 c
296.9 d
46.7 e |
0.0 a
200.1 b
236.8 c
290.0 d
45.7 e |
0.0 a
209.1 b
257.4 c
294.0 d
45.9 e |
0.0 a
47.3 e |
0.0 a
204.0 b
249.6 c
293.9 d
45.7 e |
NC
NC
NC
NC |
|
|
| CCSD=FULL |
|
|
|
|
0.0 a
204.0 b
238.6 c
285.5 d
52.8 e |
|
|
|
|
NC
NC
NC
NC |
|
0.0 a
203.2 b
249.4 c
297.6 d
47.0 e |
0.0 a
200.8 b
237.5 c
290.8 d
46.1 e |
0.0 a
212.5 b
261.0 c
296.9 d
47.6 e |
|
0.0 a
204.9 b
250.5 c
294.2 d
45.9 e |
0.0 a
213.4 b
261.9 c
296.8 d
47.8 e |
|
|
| CCSD(T) |
|
|
|
|
0.0 a
198.6 b
230.3 c
279.8 d
51.9 e |
NC
NC
NC
NC |
|
NC
NC
NC
NC |
|
|
|
0.0 a
198.6 b
243.3 c
292.8 d
46.1 e |
0.0 a
196.7 b
231.4 c
286.2 d
45.1 e |
0.0 a
204.4 b
252.1 c
288.6 d
44.8 e |
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
|
|
| CCSD(T)=FULL |
|
|
|
|
0.0 a
199.8 b
231.9 c
281.0 d
52.4 e |
|
|
|
|
|
|
0.0 a
199.3 b
244.4 c
293.5 d
46.4 e |
0.0 a
197.4 b
232.2 c
286.9 d
45.5 e |
0.0 a
208.3 b
292.0 d
46.9 e |
NC
NC |
0.0 a
201.3 b
246.2 c
289.5 d
45.5 e |
0.0 a
208.9 b
291.8 d
47.1 e |
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
cc-pVQZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
aug-cc-pVQZ |
daug-cc-pVTZ |
|---|