|
|
IV.A.4. (XIV.F.) |
Relative enthalpies of isomers - Comparison of 0K enthalpies (kJ mol-1)
Isomers of C3H4O
2015 06 30 15:39| index | Species | CAS number | Name | Relative experimental enthalpy (kJ mol-1) | sketch |
|---|---|---|---|---|---|
| a | CH2CHCHO | 107028 | Acrolein | 3.0 | 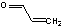 |
| b | C3H4O | 107197 | 2-Propyn-1-ol | 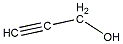 |
|
| c | C3H4O | 5009278 | Cyclopropanone | 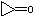 |
|
| d | C3H4O | 6004440 | Methylketene | 0.0 | 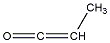 |
| e | C3H4O | 9000071 | allenol | 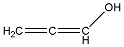 |
| composite | G3B3 | 0.0 a 119.1 b 86.2 c 0.0 d |
|---|---|---|
| G3MP2 | NC NC |
|
| G4 | NC 118.5 b 0.0 d |
|
| CBS-Q | NC NC |
| STO-3G | 3-21G | 3-21G* | 6-31G | 6-31G* | 6-31G** | 6-31+G** | 6-311G* | 6-311G** | 6-31G(2df,p) | 6-311+G(3df,2p) | TZVP | cc-pVDZ | cc-pVTZ | aug-cc-pVDZ | aug-cc-pVTZ | cc-pV(T+d)Z | daug-cc-pVTZ | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hartree fock | HF | NC NC NC |
15.1 a 130.2 b 109.9 c 0.0 d 112.0 e |
NC NC NC |
NC NC NC |
-6.4 a 148.4 b 87.9 c 0.0 d 133.2 e |
NC NC NC |
-13.5 a 123.4 b 87.0 c 0.0 d 110.3 e |
NC NC NC |
NC NC NC |
NC NC NC |
-5.0 a 128.3 b 95.6 c 0.0 d 117.1 e |
-7.9 a 121.9 b 94.0 c 0.0 d 112.9 e |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC |
NC NC NC |
-5.4 a 125.7 b 96.2 c 0.0 d |
| density functional | LSDA | 46.6 c 0.0 d 143.2 e |
82.0 c 0.0 d 126.9 e |
82.0 c 0.0 d 126.9 e |
64.7 c 0.0 d 98.8 e |
61.3 c 0.0 d 114.8 e |
61.1 c 0.0 d 104.7 e |
58.8 c 0.0 d 93.9 e |
72.2 c 0.0 d 121.5 e |
70.6 c 0.0 d 105.6 e |
65.3 c 0.0 d 107.7 e |
65.6 c 0.0 d 105.9 e |
68.6 c 0.0 d 98.1 e |
61.5 c 0.0 d 93.0 e |
68.6 c 0.0 d 98.1 e |
||||
| BLYP | NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
4.6 a 172.4 b 90.8 c 0.0 d 119.9 e |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC |
NC NC NC |
|||||
| B1B95 | 34.6 a 180.1 b 48.2 c 0.0 d |
38.1 a 177.4 b 89.8 c 0.0 d |
38.1 a 177.4 b 89.8 c 0.0 d |
7.4 a 144.8 b 72.8 c 0.0 d |
NC NC |
34.1 a 159.3 b 69.8 c 0.0 d |
13.3 a 143.9 b 68.4 c 0.0 d 103.7 e |
25.6 a 169.9 b 62.5 c 0.0 d |
24.6 a 153.8 b 77.9 c 0.0 d |
27.0 a 159.3 b 74.8 c 0.0 d |
21.5 a 162.1 b 75.1 c 0.0 d |
13.2 a 145.3 b 76.6 c 0.0 d |
6.8 a 140.7 b 70.4 c 0.0 d |
11.7 a 140.2 b 75.7 c 0.0 d |
13.2 a 145.3 b 76.6 c 0.0 d |
||||
| B3LYP | NC NC NC |
25.4 a 176.8 b 103.8 c 0.0 d 127.9 e |
NC NC NC |
NC NC NC |
3.8 a 166.8 b 84.8 c 0.0 d 121.6 e |
NC NC NC |
-3.4 a 138.6 b 82.8 c 0.0 d 99.9 e |
NC NC NC |
NC NC NC |
12.6 a 156.6 b 90.1 c 0.0 d 113.8 e |
NC NC |
3.4 a 139.3 b 91.8 c 0.0 d 103.9 e |
NC NC |
NC NC NC |
NC NC NC |
4.6 a 135.6 b 92.0 c 0.0 d 100.8 e |
NC NC NC |
||
| B3LYPultrafine | NC NC |
6.5 a 93.1 c 0.0 d |
4.6 a 135.6 b 92.0 c 0.0 d 100.7 e |
||||||||||||||||
| B3PW91 | NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC |
NC NC |
NC NC NC |
||||
| mPW1PW91 | NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC |
NC NC NC |
|||||
| M06-2X | 22.0 a 150.2 b 86.9 c 0.0 d 113.5 e |
7.8 a 151.1 b 72.2 c 0.0 d 118.6 e |
NC NC |
NC NC |
NC NC |
||||||||||||||
| PBEPBE | NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC |
NC NC NC |
NC NC NC |
NC NC |
NC NC NC |
NC NC NC |
|||
| PBEPBEultrafine | 12.1 a 74.3 c 0.0 d |
||||||||||||||||||
| PBE1PBE | 10.3 a 172.3 b 69.1 c 0.0 d 126.0 e |
||||||||||||||||||
| HSEh1PBE | 29.5 a 180.4 b 90.1 c 0.0 d |
9.8 a 172.1 b 70.5 c 0.0 d |
3.1 a 145.1 b 68.5 c 0.0 d |
12.2 a 147.2 b 77.8 c 0.0 d |
|||||||||||||||
| TPSSh | 7.5 a 176.6 b 74.5 c 0.0 d 127.9 e |
0.7 a 150.0 b 72.3 c 0.0 d 107.6 e |
15.4 a 166.0 b 79.4 c 0.0 d 120.4 e |
9.2 a 151.6 b 81.2 c 0.0 d 111.0 e |
|||||||||||||||
| wB97X-D | 29.1 a 175.6 b 91.7 c 0.0 d 131.7 e |
8.0 a 168.1 b 74.9 c 0.0 d 128.2 e |
1.5 a 141.7 b 73.4 c 0.0 d 107.4 e |
13.1 a 151.4 b 83.6 c 0.0 d 117.7 e |
8.1 a 143.1 b 81.4 c 0.0 d 112.0 e |
4.3 a 141.7 b 73.4 c 0.0 d 107.4 e |
11.6 a 145.5 b 83.6 c 0.0 d 113.2 e |
10.1 a 140.8 b 82.7 c 0.0 d 110.4 e |
|||||||||||
| B97D3 | 26.4 a 190.2 b 106.1 c 0.0 d 131.5 e |
6.1 a 177.2 b 85.2 c 0.0 d 123.9 e |
-0.4 a 149.7 b 83.0 c 0.0 d 103.4 e |
11.1 a 160.9 b 92.6 c 0.0 d 113.6 e |
7.5 a 148.8 b 90.0 c 0.0 d 106.0 e |
5.9 a 150.5 b 91.7 c 0.0 d 106.8 e |
8.9 a 152.3 b 91.8 c 0.0 d 107.1 e |
7.0 a 145.9 b 90.6 c 0.0 d 103.4 e |
|||||||||||
| STO-3G | 3-21G | 3-21G* | 6-31G | 6-31G* | 6-31G** | 6-31+G** | 6-311G* | 6-311G** | 6-31G(2df,p) | 6-311+G(3df,2p) | TZVP | cc-pVDZ | cc-pVTZ | aug-cc-pVDZ | aug-cc-pVTZ | cc-pV(T+d)Z | daug-cc-pVTZ | ||
| Moller Plesset perturbation | MP2 | NC NC NC |
25.8 a 147.6 b 116.9 c 0.0 d 147.2 e |
NC NC NC |
NC NC NC |
2.7 a 144.2 b 79.1 c 0.0 d 135.8 e |
NC NC NC |
NC NC |
11.7 a 152.1 b 87.9 c 0.0 d 145.8 e |
NC NC NC |
NC NC NC |
3.2 a 87.4 c 0.0 d 120.5 e |
NC NC NC |
7.9 a 123.3 b 86.2 c 0.0 d |
NC NC NC |
6.1 a 117.1 b 85.3 c 0.0 d |
NC NC |
||
| MP2=FULL | NC NC NC |
NC NC NC |
NC NC |
NC NC |
3.9 a 145.1 b 79.4 c 0.0 d 136.4 e |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
NC NC NC |
9.2 a 125.3 b 86.0 c 0.0 d |
NC NC |
||||||
| MP3 | NC NC |
NC NC |
|||||||||||||||||
| MP3=FULL | -7.1 a 137.3 b 73.7 c 0.0 d 114.7 e |
-13.2 a 117.5 b 73.8 c 0.0 d 97.7 e |
|||||||||||||||||
| MP4 | NC NC |
||||||||||||||||||
| B2PLYP | 3.1 a 158.9 b 84.6 c 0.0 d 129.0 e |
NC NC |
NC NC |
NC NC |
6.1 a 135.2 b 92.3 c 0.0 d 110.6 e |
NC NC |
|||||||||||||
| B2PLYP=FULL | NC NC |
NC NC |
NC NC |
||||||||||||||||
| B2PLYP=FULLultrafine | 10.1 a 0.0 d |
||||||||||||||||||
| Configuration interaction | CID | NC NC |
NC NC |
NC NC |
NC NC NC |
NC NC |
|||||||||||||
| CISD | NC NC |
NC NC |
NC NC |
NC NC NC |
NC NC |
||||||||||||||
| STO-3G | 3-21G | 3-21G* | 6-31G | 6-31G* | 6-31G** | 6-31+G** | 6-311G* | 6-311G** | 6-31G(2df,p) | 6-311+G(3df,2p) | TZVP | cc-pVDZ | cc-pVTZ | aug-cc-pVDZ | aug-cc-pVTZ | cc-pV(T+d)Z | daug-cc-pVTZ | ||
| Quadratic configuration interaction | QCISD | NC NC |
NC NC NC |
NC NC |
NC NC |
NC NC NC |
NC NC |
NC NC |
NC NC NC |
NC NC |
NC NC |
NC NC |
NC NC |
||||||
| Coupled Cluster | CCD | NC NC |
NC NC |
NC NC |
NC NC |
-8.1 a 135.1 b 74.6 c 0.0 d 115.5 e |
NC NC |
NC NC |
NC NC |
NC NC |
NC NC |
||||||||
| CCSD | NC NC |
NC NC |
|||||||||||||||||
| CCSD=FULL | NC NC |
NC NC |
NC NC |
||||||||||||||||
| CCSD(T) | NC NC |
||||||||||||||||||
| STO-3G | 3-21G | 3-21G* | 6-31G | 6-31G* | 6-31G** | 6-31+G** | 6-311G* | 6-311G** | 6-31G(2df,p) | 6-311+G(3df,2p) | TZVP | cc-pVDZ | cc-pVTZ | aug-cc-pVDZ | aug-cc-pVTZ | cc-pV(T+d)Z | daug-cc-pVTZ |
| CEP-31G | CEP-31G* | CEP-121G | CEP-121G* | LANL2DZ | SDD | cc-pVTZ-PP | aug-cc-pVTZ-PP | Def2TZVPP | ||
|---|---|---|---|---|---|---|---|---|---|---|
| hartree fock | HF | -90046.0 a -89930.8 c -89916.2 e |
-90277.9 a -90182.9 c -90116.1 e |
-90071.3 a -89954.2 c -89963.5 e |
-90313.5 a -90212.0 c -90176.6 e |
NC NC NC |
NC NC NC |
-4.5 a 127.2 b 96.7 c 0.0 d 115.6 e |
||
| density functional | B1B95 | -2.5 a 142.4 b 53.2 c 0.0 d 99.6 e |
1.2 a 174.0 b 51.5 c 0.0 d 127.8 e |
|||||||
| B3LYP | -92350.2 a -92256.9 c -92230.8 e |
-92494.5 a -92415.4 c -92360.3 e |
-92380.4 a -92279.2 c -92276.6 e |
-92535.0 a -92447.0 c -92420.0 e |
NC NC NC |
NC NC NC |
5.9 a 139.4 b 92.5 c 0.0 d 103.2 e |
|||
| PBEPBE | 13.8 a 151.3 b 79.9 c 0.0 d 105.1 e |
|||||||||
| Moller Plesset perturbation | MP2 | -90960.5 a -90855.2 c -90822.6 e |
-91654.2 a -91579.9 c -91502.0 e |
-91092.9 a -90980.7 c -90969.6 e |
-91785.8 a -91703.5 c -91654.4 e |
NC NC |
NC NC NC |
7.8 a 121.1 b 85.6 c 0.0 d 114.8 e |
| 6-311+G(3df,2p) | cc-pVDZ | cc-pVTZ | aug-cc-pVDZ | aug-cc-pVTZ | cc-pV(T+d)Z | ||
|---|---|---|---|---|---|---|---|
| Moller Plesset perturbation | MP2FC// HF/6-31G* | 126.6 b 0.0 d |
128.7 b 0.0 d |
128.7 b 87.3 c 0.0 d |
|||
| MP2FC// B3LYP/6-31G* | 84.9 c 0.0 d |
147.6 b 0.0 d |
123.4 b 0.0 d |
123.4 b 86.5 c 0.0 d |
|||
| MP2FC// MP2FC/6-31G* | 86.8 c 0.0 d |
86.3 c 0.0 d |
86.8 c 0.0 d |
||||
| MP4// HF/6-31G* | 2010.6 a 134.6 b 0.0 d |
2014.0 a 136.2 b 0.0 d |
136.2 b 89.8 c 0.0 d |
||||
| MP4// B3LYP/6-31G* | 1662.2 a 157.9 b 0.0 d |
2025.5 a 132.6 b 0.0 d |
132.6 b 89.4 c 0.0 d |
||||
| MP4// MP2/6-31G* | 2028.4 a 0.0 d |
90.2 c 0.0 d |
|||||
| Coupled Cluster | CCSD// B3LYP/6-31G* | 82.0 c 0.0 d |
|||||
| CCSD// MP2FC/6-31G* | 83.2 c 0.0 d |
83.3 c 0.0 d |
|||||
| CCSD(T)// MP2FC/6-31G* | 85.2 c 0.0 d |
85.2 c 0.0 d |
For descriptions of the methods (AM1, HF, MP2, ...) and basis sets (3-21G, 3-21G*, 6-31G, ...) see the glossary in section I.C. Predefined means the basis set used is determined by the method.
gaw refers to the group additivity method implemeted in the NIST Chemistry Webbook.
See section III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.