Vibrational Frequencies calculated at BLYP/6-31+G**
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3450 |
3431 |
0.38 |
|
|
|
| 2 |
A |
3362 |
3344 |
4.79 |
|
|
|
| 3 |
A |
2999 |
2983 |
47.12 |
|
|
|
| 4 |
A |
2985 |
2969 |
97.26 |
|
|
|
| 5 |
A |
2980 |
2964 |
53.58 |
|
|
|
| 6 |
A |
2978 |
2962 |
64.40 |
|
|
|
| 7 |
A |
2968 |
2952 |
74.30 |
|
|
|
| 8 |
A |
2939 |
2923 |
80.90 |
|
|
|
| 9 |
A |
2934 |
2919 |
13.99 |
|
|
|
| 10 |
A |
2932 |
2916 |
34.66 |
|
|
|
| 11 |
A |
2930 |
2914 |
17.59 |
|
|
|
| 12 |
A |
2918 |
2903 |
18.35 |
|
|
|
| 13 |
A |
2916 |
2900 |
9.39 |
|
|
|
| 14 |
A |
1616 |
1607 |
31.67 |
|
|
|
| 15 |
A |
1472 |
1464 |
1.42 |
|
|
|
| 16 |
A |
1459 |
1451 |
10.90 |
|
|
|
| 17 |
A |
1454 |
1446 |
3.25 |
|
|
|
| 18 |
A |
1452 |
1444 |
0.99 |
|
|
|
| 19 |
A |
1445 |
1438 |
0.29 |
|
|
|
| 20 |
A |
1374 |
1367 |
3.86 |
|
|
|
| 21 |
A |
1337 |
1330 |
4.41 |
|
|
|
| 22 |
A |
1333 |
1326 |
0.59 |
|
|
|
| 23 |
A |
1331 |
1324 |
3.21 |
|
|
|
| 24 |
A |
1324 |
1317 |
3.38 |
|
|
|
| 25 |
A |
1322 |
1315 |
0.46 |
|
|
|
| 26 |
A |
1294 |
1287 |
0.61 |
|
|
|
| 27 |
A |
1262 |
1255 |
1.39 |
|
|
|
| 28 |
A |
1247 |
1241 |
2.83 |
|
|
|
| 29 |
A |
1238 |
1231 |
1.07 |
|
|
|
| 30 |
A |
1186 |
1179 |
3.14 |
|
|
|
| 31 |
A |
1157 |
1151 |
0.95 |
|
|
|
| 32 |
A |
1077 |
1071 |
9.35 |
|
|
|
| 33 |
A |
1069 |
1063 |
5.50 |
|
|
|
| 34 |
A |
1052 |
1047 |
0.53 |
|
|
|
| 35 |
A |
1037 |
1031 |
0.58 |
|
|
|
| 36 |
A |
1005 |
999 |
7.03 |
|
|
|
| 37 |
A |
997 |
992 |
2.97 |
|
|
|
| 38 |
A |
949 |
944 |
1.94 |
|
|
|
| 39 |
A |
901 |
896 |
1.95 |
|
|
|
| 40 |
A |
874 |
869 |
41.00 |
|
|
|
| 41 |
A |
855 |
850 |
3.28 |
|
|
|
| 42 |
A |
821 |
817 |
66.51 |
|
|
|
| 43 |
A |
813 |
808 |
58.24 |
|
|
|
| 44 |
A |
768 |
764 |
0.60 |
|
|
|
| 45 |
A |
752 |
748 |
4.50 |
|
|
|
| 46 |
A |
536 |
533 |
1.02 |
|
|
|
| 47 |
A |
448 |
445 |
1.33 |
|
|
|
| 48 |
A |
436 |
434 |
2.10 |
|
|
|
| 49 |
A |
394 |
392 |
0.20 |
|
|
|
| 50 |
A |
332 |
330 |
12.61 |
|
|
|
| 51 |
A |
322 |
321 |
1.18 |
|
|
|
| 52 |
A |
246 |
245 |
28.61 |
|
|
|
| 53 |
A |
218 |
216 |
12.66 |
|
|
|
| 54 |
A |
154 |
153 |
0.31 |
|
|
|
Unscaled Zero Point Vibrational Energy (zpe) 39823.2 cm
-1
Scaled (by 0.9947) Zero Point Vibrational Energy (zpe) 39612.1 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Charges, Dipole, Quadrupole and Polarizability
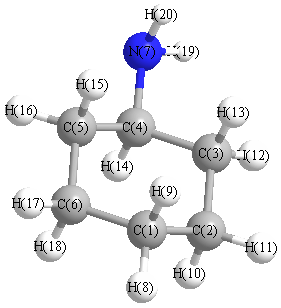
Charges from optimized geometry at BLYP/6-31+G**
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.272 |
|
|
|
| 2 |
C |
-0.277 |
|
|
|
| 3 |
C |
-0.252 |
|
|
|
| 4 |
C |
-0.089 |
|
|
|
| 5 |
C |
-0.238 |
|
|
|
| 6 |
C |
-0.279 |
|
|
|
| 7 |
N |
-0.571 |
|
|
|
| 8 |
H |
0.129 |
|
|
|
| 9 |
H |
0.132 |
|
|
|
| 10 |
H |
0.133 |
|
|
|
| 11 |
H |
0.130 |
|
|
|
| 12 |
H |
0.146 |
|
|
|
| 13 |
H |
0.124 |
|
|
|
| 14 |
H |
0.123 |
|
|
|
| 15 |
H |
0.130 |
|
|
|
| 16 |
H |
0.124 |
|
|
|
| 17 |
H |
0.129 |
|
|
|
| 18 |
H |
0.133 |
|
|
|
| 19 |
H |
0.273 |
|
|
|
| 20 |
H |
0.271 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.727 |
1.105 |
0.436 |
1.393 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-49.809 |
-4.183 |
-1.589 |
| y |
-4.183 |
-47.829 |
-0.813 |
| z |
-1.589 |
-0.813 |
-45.681 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
-3.054 |
-4.183 |
-1.589 |
| y |
-4.183 |
-0.084 |
-0.813 |
| z |
-1.589 |
-0.813 |
3.138 |
|
| Polar |
|---|
| 3z2-r2 | 6.276 |
|---|
| x2-y2 | -1.980 |
|---|
| xy | -4.183 |
|---|
| xz | -1.589 |
|---|
| yz | -0.813 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
13.535 |
0.043 |
-0.089 |
| y |
0.043 |
12.434 |
0.059 |
| z |
-0.089 |
0.059 |
10.740 |
<r2> (average value of r
2) Å
2
| <r2> |
239.165 |
| (<r2>)1/2 |
15.465 |