Jump to
S1C2
Vibrational Frequencies calculated at B2PLYP/6-311+G(3df,2p)
Geometric Data calculated at B2PLYP/6-311+G(3df,2p)
Electronic energy levels
Charges, Dipole, Quadrupole and Polarizability
Jump to
S1C1
Energy calculated at B2PLYP/6-311+G(3df,2p)
| | hartrees |
|---|
| Energy at 0K | -194.253948 |
| Energy at 298.15K | |
| HF Energy | -194.004404 |
| Nuclear repulsion energy | 132.804806 |
The energy at 298.15K was derived from the energy at 0K
and an integrated heat capacity that used the calculated vibrational frequencies.
Vibrational Frequencies calculated at B2PLYP/6-311+G(3df,2p)
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3874 |
3874 |
34.55 |
104.45 |
0.23 |
0.38 |
| 2 |
A |
3136 |
3136 |
22.53 |
33.88 |
0.74 |
0.85 |
| 3 |
A |
3113 |
3113 |
44.56 |
55.74 |
0.55 |
0.71 |
| 4 |
A |
3082 |
3082 |
20.79 |
74.28 |
0.71 |
0.83 |
| 5 |
A |
3049 |
3049 |
53.87 |
158.23 |
0.06 |
0.11 |
| 6 |
A |
3047 |
3047 |
35.03 |
125.86 |
0.04 |
0.08 |
| 7 |
A |
3033 |
3033 |
14.64 |
95.02 |
0.57 |
0.73 |
| 8 |
A |
3000 |
3000 |
57.31 |
115.06 |
0.09 |
0.16 |
| 9 |
A |
1534 |
1534 |
2.75 |
3.29 |
0.75 |
0.86 |
| 10 |
A |
1524 |
1524 |
7.05 |
2.20 |
0.75 |
0.86 |
| 11 |
A |
1511 |
1511 |
7.22 |
6.42 |
0.75 |
0.86 |
| 12 |
A |
1492 |
1492 |
2.68 |
6.72 |
0.72 |
0.84 |
| 13 |
A |
1455 |
1455 |
4.92 |
0.99 |
0.16 |
0.27 |
| 14 |
A |
1421 |
1421 |
2.69 |
0.13 |
0.75 |
0.86 |
| 15 |
A |
1389 |
1389 |
1.28 |
0.80 |
0.74 |
0.85 |
| 16 |
A |
1330 |
1330 |
14.90 |
4.90 |
0.71 |
0.83 |
| 17 |
A |
1280 |
1280 |
0.34 |
2.95 |
0.73 |
0.85 |
| 18 |
A |
1248 |
1248 |
36.87 |
1.86 |
0.35 |
0.52 |
| 19 |
A |
1169 |
1169 |
5.17 |
1.14 |
0.08 |
0.15 |
| 20 |
A |
1121 |
1121 |
10.18 |
3.69 |
0.48 |
0.65 |
| 21 |
A |
1073 |
1073 |
43.07 |
2.96 |
0.59 |
0.74 |
| 22 |
A |
988 |
988 |
50.49 |
3.47 |
0.69 |
0.81 |
| 23 |
A |
932 |
932 |
2.18 |
0.40 |
0.71 |
0.83 |
| 24 |
A |
872 |
872 |
1.39 |
11.88 |
0.09 |
0.16 |
| 25 |
A |
778 |
778 |
0.86 |
0.41 |
0.25 |
0.40 |
| 26 |
A |
478 |
478 |
8.28 |
0.23 |
0.48 |
0.65 |
| 27 |
A |
327 |
327 |
5.54 |
0.29 |
0.29 |
0.45 |
| 28 |
A |
245 |
245 |
104.67 |
1.49 |
0.74 |
0.85 |
| 29 |
A |
225 |
225 |
3.59 |
0.05 |
0.48 |
0.65 |
| 30 |
A |
142 |
142 |
9.46 |
0.14 |
0.73 |
0.85 |
Unscaled Zero Point Vibrational Energy (zpe) 23934.0 cm
-1
Scaled (by 1) Zero Point Vibrational Energy (zpe) 23934.0 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Geometric Data calculated at B2PLYP/6-311+G(3df,2p)
Point Group is C1
Cartesians (Å)
| Atom |
x (Å) |
y (Å) |
z (Å) |
|---|
| C1 |
-1.539 |
-0.515 |
0.128 |
| C2 |
-0.633 |
0.638 |
-0.292 |
| C3 |
0.762 |
0.545 |
0.296 |
| O4 |
1.385 |
-0.632 |
-0.216 |
| H5 |
-2.531 |
-0.414 |
-0.312 |
| H6 |
-1.658 |
-0.542 |
1.213 |
| H7 |
-1.123 |
-1.469 |
-0.187 |
| H8 |
-0.546 |
0.670 |
-1.380 |
| H9 |
-1.067 |
1.591 |
0.019 |
| H10 |
0.704 |
0.499 |
1.389 |
| H11 |
1.343 |
1.431 |
0.024 |
| H12 |
2.258 |
-0.719 |
0.172 |
Atom - Atom Distances (Å)
| |
C1 |
C2 |
C3 |
O4 |
H5 |
H6 |
H7 |
H8 |
H9 |
H10 |
H11 |
H12 |
| C1 | | 1.5259 | 2.5389 | 2.9472 | 1.0893 | 1.0910 | 1.0877 | 2.1600 | 2.1612 | 2.7655 | 3.4791 | 3.8030 |
C2 | 1.5259 | | 1.5166 | 2.3860 | 2.1702 | 2.1697 | 2.1661 | 1.0916 | 1.0923 | 2.1521 | 2.1521 | 3.2271 | C3 | 2.5389 | 1.5166 | | 1.4266 | 3.4827 | 2.8064 | 2.8000 | 2.1294 | 2.1249 | 1.0956 | 1.0941 | 1.9621 | O4 | 2.9472 | 2.3860 | 1.4266 | | 3.9232 | 3.3629 | 2.6444 | 2.6040 | 3.3182 | 2.0785 | 2.0772 | 0.9589 | H5 | 1.0893 | 2.1702 | 3.4827 | 3.9232 | | 1.7613 | 1.7633 | 2.5010 | 2.5050 | 3.7665 | 4.3036 | 4.8226 | H6 | 1.0910 | 2.1697 | 2.8064 | 3.3629 | 1.7613 | | 1.7618 | 3.0701 | 2.5147 | 2.5867 | 3.7828 | 4.0554 | H7 | 1.0877 | 2.1661 | 2.8000 | 2.6444 | 1.7633 | 1.7618 | | 2.5162 | 3.0676 | 3.1134 | 3.8123 | 3.4817 | H8 | 2.1600 | 1.0916 | 2.1294 | 2.6040 | 2.5010 | 3.0701 | 2.5162 | | 1.7543 | 3.0425 | 2.4732 | 3.4928 | H9 | 2.1612 | 1.0923 | 2.1249 | 3.3182 | 2.5050 | 2.5147 | 3.0676 | 1.7543 | | 2.4903 | 2.4149 | 4.0512 | H10 | 2.7655 | 2.1521 | 1.0956 | 2.0785 | 3.7665 | 2.5867 | 3.1134 | 3.0425 | 2.4903 | | 1.7722 | 2.3197 | H11 | 3.4791 | 2.1521 | 1.0941 | 2.0772 | 4.3036 | 3.7828 | 3.8123 | 2.4732 | 2.4149 | 1.7722 | | 2.3410 | H12 | 3.8030 | 3.2271 | 1.9621 | 0.9589 | 4.8226 | 4.0554 | 3.4817 | 3.4928 | 4.0512 | 2.3197 | 2.3410 | |
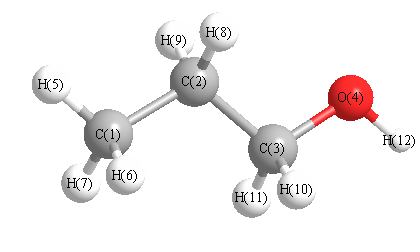 More geometry information
More geometry information
Calculated Bond Angles
| atom1 |
atom2 |
atom3 |
angle |
|
atom1 |
atom2 |
atom3 |
angle |
| C1 |
C2 |
C3 |
113.127 |
|
C1 |
C2 |
H8 |
110.112 |
| C1 |
C2 |
H9 |
110.167 |
|
C2 |
C1 |
H5 |
111.067 |
| C2 |
C1 |
H6 |
110.925 |
|
C2 |
C1 |
H7 |
110.832 |
| C2 |
C3 |
O4 |
108.291 |
|
C2 |
C3 |
H10 |
109.901 |
| C2 |
C3 |
H11 |
109.991 |
|
C3 |
C2 |
H8 |
108.357 |
| C3 |
C2 |
H9 |
107.971 |
|
C3 |
O4 |
H12 |
109.099 |
| O4 |
C3 |
H10 |
110.308 |
|
O4 |
C3 |
H11 |
110.291 |
| H5 |
C1 |
H6 |
107.770 |
|
H5 |
C1 |
H7 |
108.191 |
| H6 |
C1 |
H7 |
107.924 |
|
H8 |
C2 |
H9 |
106.884 |
| H10 |
C3 |
H11 |
108.059 |
|
Electronic energy levels
Charges, Dipole, Quadrupole and Polarizability
Charges from optimized geometry at B2PLYP/6-311+G(3df,2p)
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.473 |
|
|
|
| 2 |
C |
-0.186 |
|
|
|
| 3 |
C |
0.056 |
|
|
|
| 4 |
O |
-0.622 |
|
|
|
| 5 |
H |
0.138 |
|
|
|
| 6 |
H |
0.146 |
|
|
|
| 7 |
H |
0.162 |
|
|
|
| 8 |
H |
0.151 |
|
|
|
| 9 |
H |
0.138 |
|
|
|
| 10 |
H |
0.140 |
|
|
|
| 11 |
H |
0.129 |
|
|
|
| 12 |
H |
0.221 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.614 |
1.005 |
0.986 |
1.536 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-23.438 |
-0.076 |
2.038 |
| y |
-0.076 |
-27.694 |
-0.567 |
| z |
2.038 |
-0.567 |
-27.273 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
4.045 |
-0.076 |
2.038 |
| y |
-0.076 |
-2.338 |
-0.567 |
| z |
2.038 |
-0.567 |
-1.706 |
|
| Polar |
|---|
| 3z2-r2 | -3.413 |
|---|
| x2-y2 | 4.256 |
|---|
| xy | -0.076 |
|---|
| xz | 2.038 |
|---|
| yz | -0.567 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
7.153 |
0.067 |
0.064 |
| y |
0.067 |
6.462 |
-0.015 |
| z |
0.064 |
-0.015 |
6.050 |
<r2> (average value of r
2) Å
2
| <r2> |
95.430 |
| (<r2>)1/2 |
9.769 |
