Jump to
S1C2
Energy calculated at M06-2X/6-311+G(3df,2p)
| | hartrees |
|---|
| Energy at 0K | -194.328267 |
| Energy at 298.15K | |
| HF Energy | -194.328267 |
| Nuclear repulsion energy | 130.891164 |
The energy at 298.15K was derived from the energy at 0K
and an integrated heat capacity that used the calculated vibrational frequencies.
Vibrational Frequencies calculated at M06-2X/6-311+G(3df,2p)
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A' |
3919 |
3919 |
43.19 |
113.65 |
0.24 |
0.39 |
| 2 |
A' |
3133 |
3133 |
23.84 |
68.11 |
0.56 |
0.72 |
| 3 |
A' |
3063 |
3063 |
28.67 |
53.67 |
0.13 |
0.23 |
| 4 |
A' |
3053 |
3053 |
16.89 |
193.30 |
0.01 |
0.01 |
| 5 |
A' |
3002 |
3002 |
47.62 |
111.92 |
0.05 |
0.09 |
| 6 |
A' |
1533 |
1533 |
5.24 |
2.81 |
0.75 |
0.86 |
| 7 |
A' |
1512 |
1512 |
5.55 |
0.18 |
0.47 |
0.64 |
| 8 |
A' |
1498 |
1498 |
0.87 |
10.15 |
0.73 |
0.84 |
| 9 |
A' |
1459 |
1459 |
3.04 |
0.72 |
0.44 |
0.61 |
| 10 |
A' |
1415 |
1415 |
1.58 |
0.10 |
0.25 |
0.40 |
| 11 |
A' |
1347 |
1347 |
8.88 |
0.38 |
0.40 |
0.57 |
| 12 |
A' |
1258 |
1258 |
49.83 |
1.75 |
0.72 |
0.84 |
| 13 |
A' |
1123 |
1123 |
49.40 |
8.53 |
0.37 |
0.54 |
| 14 |
A' |
1085 |
1085 |
58.46 |
2.78 |
0.12 |
0.22 |
| 15 |
A' |
1059 |
1059 |
2.98 |
2.92 |
0.47 |
0.64 |
| 16 |
A' |
897 |
897 |
5.35 |
7.86 |
0.16 |
0.28 |
| 17 |
A' |
464 |
464 |
10.58 |
2.13 |
0.21 |
0.35 |
| 18 |
A' |
277 |
277 |
3.98 |
0.07 |
0.23 |
0.38 |
| 19 |
A" |
3122 |
3122 |
48.84 |
6.14 |
0.75 |
0.86 |
| 20 |
A" |
3098 |
3098 |
2.64 |
77.62 |
0.75 |
0.86 |
| 21 |
A" |
3033 |
3033 |
32.85 |
76.42 |
0.75 |
0.86 |
| 22 |
A" |
1506 |
1506 |
7.78 |
4.28 |
0.75 |
0.86 |
| 23 |
A" |
1323 |
1323 |
0.07 |
6.60 |
0.75 |
0.86 |
| 24 |
A" |
1272 |
1272 |
0.19 |
0.11 |
0.75 |
0.86 |
| 25 |
A" |
1189 |
1189 |
0.99 |
0.35 |
0.75 |
0.86 |
| 26 |
A" |
901 |
901 |
1.89 |
0.05 |
0.75 |
0.86 |
| 27 |
A" |
762 |
762 |
0.96 |
0.09 |
0.75 |
0.86 |
| 28 |
A" |
273 |
273 |
112.62 |
1.53 |
0.75 |
0.86 |
| 29 |
A" |
232 |
232 |
2.55 |
0.05 |
0.75 |
0.86 |
| 30 |
A" |
128 |
128 |
5.71 |
0.03 |
0.75 |
0.86 |
Unscaled Zero Point Vibrational Energy (zpe) 23967.1 cm
-1
Scaled (by 1) Zero Point Vibrational Energy (zpe) 23967.1 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Geometric Data calculated at M06-2X/6-311+G(3df,2p)
Point Group is Cs
Cartesians (Å)
| Atom |
x (Å) |
y (Å) |
z (Å) |
|---|
| C1 |
-1.450 |
1.211 |
0.000 |
| C2 |
0.000 |
0.745 |
0.000 |
| C3 |
0.101 |
-0.766 |
0.000 |
| O4 |
1.472 |
-1.121 |
0.000 |
| H5 |
-1.515 |
2.298 |
0.000 |
| H6 |
-1.980 |
0.847 |
0.881 |
| H7 |
-1.980 |
0.847 |
-0.881 |
| H8 |
0.526 |
1.126 |
0.877 |
| H9 |
0.526 |
1.126 |
-0.877 |
| H10 |
-0.404 |
-1.170 |
0.886 |
| H11 |
-0.404 |
-1.170 |
-0.886 |
| H12 |
1.554 |
-2.075 |
0.000 |
Atom - Atom Distances (Å)
| |
C1 |
C2 |
C3 |
O4 |
H5 |
H6 |
H7 |
H8 |
H9 |
H10 |
H11 |
H12 |
| C1 | | 1.5238 | 2.5138 | 3.7388 | 1.0889 | 1.0912 | 1.0912 | 2.1639 | 2.1639 | 2.7479 | 2.7479 | 4.4532 |
C2 | 1.5238 | | 1.5144 | 2.3760 | 2.1700 | 2.1700 | 2.1700 | 1.0916 | 1.0916 | 2.1478 | 2.1478 | 3.2198 | C3 | 2.5138 | 1.5144 | | 1.4158 | 3.4647 | 2.7769 | 2.7769 | 2.1289 | 2.1289 | 1.0965 | 1.0965 | 1.9557 | O4 | 3.7388 | 2.3760 | 1.4158 | | 4.5398 | 4.0700 | 4.0700 | 2.5909 | 2.5909 | 2.0750 | 2.0750 | 0.9580 | H5 | 1.0889 | 2.1700 | 3.4647 | 4.5398 | | 1.7611 | 1.7611 | 2.5114 | 2.5114 | 3.7481 | 3.7481 | 5.3430 | H6 | 1.0912 | 2.1700 | 2.7769 | 4.0700 | 1.7611 | | 1.7628 | 2.5218 | 3.0743 | 2.5597 | 3.1104 | 4.6699 | H7 | 1.0912 | 2.1700 | 2.7769 | 4.0700 | 1.7611 | 1.7628 | | 3.0743 | 2.5218 | 3.1104 | 2.5597 | 4.6699 | H8 | 2.1639 | 1.0916 | 2.1289 | 2.5909 | 2.5114 | 2.5218 | 3.0743 | | 1.7541 | 2.4774 | 3.0406 | 3.4751 | H9 | 2.1639 | 1.0916 | 2.1289 | 2.5909 | 2.5114 | 3.0743 | 2.5218 | 1.7541 | | 3.0406 | 2.4774 | 3.4751 | H10 | 2.7479 | 2.1478 | 1.0965 | 2.0750 | 3.7481 | 2.5597 | 3.1104 | 2.4774 | 3.0406 | | 1.7715 | 2.3322 | H11 | 2.7479 | 2.1478 | 1.0965 | 2.0750 | 3.7481 | 3.1104 | 2.5597 | 3.0406 | 2.4774 | 1.7715 | | 2.3322 | H12 | 4.4532 | 3.2198 | 1.9557 | 0.9580 | 5.3430 | 4.6699 | 4.6699 | 3.4751 | 3.4751 | 2.3322 | 2.3322 | |
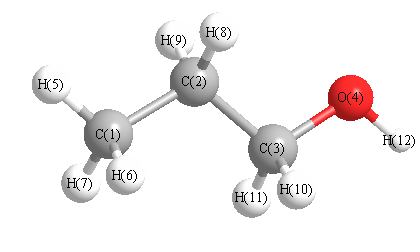 More geometry information
More geometry information
Calculated Bond Angles
| atom1 |
atom2 |
atom3 |
angle |
|
atom1 |
atom2 |
atom3 |
angle |
| C1 |
C2 |
C3 |
111.669 |
|
C1 |
C2 |
H8 |
110.576 |
| C1 |
C2 |
H9 |
110.576 |
|
C2 |
C1 |
H5 |
111.221 |
| C2 |
C1 |
H6 |
111.086 |
|
C2 |
C1 |
H7 |
111.086 |
| C2 |
C3 |
O4 |
108.315 |
|
C2 |
C3 |
H10 |
109.660 |
| C2 |
C3 |
H11 |
109.660 |
|
C3 |
C2 |
H8 |
108.472 |
| C3 |
C2 |
H9 |
108.472 |
|
C3 |
O4 |
H12 |
109.435 |
| O4 |
C3 |
H10 |
110.720 |
|
O4 |
C3 |
H11 |
110.720 |
| H5 |
C1 |
H6 |
107.766 |
|
H5 |
C1 |
H7 |
107.766 |
| H6 |
C1 |
H7 |
107.753 |
|
H8 |
C2 |
H9 |
106.923 |
| H10 |
C3 |
H11 |
107.759 |
|
Electronic energy levels
Charges, Dipole, Quadrupole and Polarizability
Charges from optimized geometry at M06-2X/6-311+G(3df,2p)
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.625 |
|
|
|
| 2 |
C |
-0.267 |
|
|
|
| 3 |
C |
-0.119 |
|
|
|
| 4 |
O |
-0.628 |
|
|
|
| 5 |
H |
0.185 |
|
|
|
| 6 |
H |
0.201 |
|
|
|
| 7 |
H |
0.201 |
|
|
|
| 8 |
H |
0.209 |
|
|
|
| 9 |
H |
0.209 |
|
|
|
| 10 |
H |
0.201 |
|
|
|
| 11 |
H |
0.201 |
|
|
|
| 12 |
H |
0.232 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
-1.206 |
-0.911 |
0.000 |
1.512 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-29.935 |
-0.955 |
0.000 |
| y |
-0.955 |
-22.111 |
0.000 |
| z |
0.000 |
0.000 |
-26.860 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
-5.449 |
-0.955 |
0.000 |
| y |
-0.955 |
6.286 |
0.000 |
| z |
0.000 |
0.000 |
-0.837 |
|
| Polar |
|---|
| 3z2-r2 | -1.673 |
|---|
| x2-y2 | -7.824 |
|---|
| xy | -0.955 |
|---|
| xz | 0.000 |
|---|
| yz | 0.000 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
6.854 |
-0.646 |
0.000 |
| y |
-0.646 |
6.793 |
0.000 |
| z |
0.000 |
0.000 |
5.754 |
<r2> (average value of r
2) Å
2
| <r2> |
106.538 |
| (<r2>)1/2 |
10.322 |
Jump to
S1C1
Energy calculated at M06-2X/6-311+G(3df,2p)
| | hartrees |
|---|
| Energy at 0K | -194.328536 |
| Energy at 298.15K | |
| HF Energy | -194.328536 |
| Nuclear repulsion energy | 133.223457 |
The energy at 298.15K was derived from the energy at 0K
and an integrated heat capacity that used the calculated vibrational frequencies.
Vibrational Frequencies calculated at M06-2X/6-311+G(3df,2p)
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3922 |
3922 |
46.29 |
101.99 |
0.24 |
0.39 |
| 2 |
A |
3144 |
3144 |
17.10 |
33.78 |
0.75 |
0.86 |
| 3 |
A |
3124 |
3124 |
35.61 |
49.91 |
0.52 |
0.69 |
| 4 |
A |
3094 |
3094 |
15.89 |
74.69 |
0.69 |
0.82 |
| 5 |
A |
3060 |
3060 |
57.82 |
79.10 |
0.11 |
0.19 |
| 6 |
A |
3056 |
3056 |
12.18 |
185.69 |
0.01 |
0.01 |
| 7 |
A |
3041 |
3041 |
21.75 |
83.70 |
0.67 |
0.80 |
| 8 |
A |
3006 |
3006 |
52.13 |
114.56 |
0.08 |
0.15 |
| 9 |
A |
1530 |
1530 |
3.47 |
3.16 |
0.75 |
0.86 |
| 10 |
A |
1513 |
1513 |
8.01 |
1.98 |
0.75 |
0.86 |
| 11 |
A |
1500 |
1500 |
7.49 |
6.08 |
0.75 |
0.86 |
| 12 |
A |
1483 |
1483 |
2.81 |
6.60 |
0.72 |
0.84 |
| 13 |
A |
1454 |
1454 |
4.77 |
0.94 |
0.11 |
0.19 |
| 14 |
A |
1413 |
1413 |
3.69 |
0.23 |
0.71 |
0.83 |
| 15 |
A |
1378 |
1378 |
0.83 |
0.87 |
0.74 |
0.85 |
| 16 |
A |
1323 |
1323 |
20.13 |
5.04 |
0.71 |
0.83 |
| 17 |
A |
1279 |
1279 |
1.71 |
2.82 |
0.73 |
0.85 |
| 18 |
A |
1243 |
1243 |
40.32 |
1.56 |
0.41 |
0.58 |
| 19 |
A |
1167 |
1167 |
9.28 |
1.00 |
0.09 |
0.17 |
| 20 |
A |
1142 |
1142 |
22.44 |
4.30 |
0.43 |
0.60 |
| 21 |
A |
1079 |
1079 |
37.79 |
2.74 |
0.63 |
0.77 |
| 22 |
A |
1002 |
1002 |
34.53 |
3.09 |
0.72 |
0.84 |
| 23 |
A |
927 |
927 |
1.73 |
0.28 |
0.52 |
0.68 |
| 24 |
A |
881 |
881 |
2.32 |
11.98 |
0.09 |
0.17 |
| 25 |
A |
777 |
777 |
0.52 |
0.31 |
0.32 |
0.49 |
| 26 |
A |
481 |
481 |
7.95 |
0.21 |
0.49 |
0.65 |
| 27 |
A |
330 |
330 |
4.02 |
0.26 |
0.26 |
0.41 |
| 28 |
A |
233 |
233 |
23.96 |
0.60 |
0.70 |
0.82 |
| 29 |
A |
223 |
223 |
83.64 |
1.23 |
0.75 |
0.86 |
| 30 |
A |
140 |
140 |
13.47 |
0.11 |
0.74 |
0.85 |
Unscaled Zero Point Vibrational Energy (zpe) 23973.2 cm
-1
Scaled (by 1) Zero Point Vibrational Energy (zpe) 23973.2 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Geometric Data calculated at M06-2X/6-311+G(3df,2p)
Point Group is C1
Cartesians (Å)
| Atom |
x (Å) |
y (Å) |
z (Å) |
|---|
| C1 |
-1.524 |
-0.519 |
0.128 |
| C2 |
-0.634 |
0.646 |
-0.290 |
| C3 |
0.761 |
0.543 |
0.295 |
| O4 |
1.369 |
-0.630 |
-0.218 |
| H5 |
-2.519 |
-0.431 |
-0.307 |
| H6 |
-1.633 |
-0.552 |
1.213 |
| H7 |
-1.091 |
-1.464 |
-0.196 |
| H8 |
-0.548 |
0.683 |
-1.378 |
| H9 |
-1.072 |
1.593 |
0.032 |
| H10 |
0.700 |
0.496 |
1.389 |
| H11 |
1.348 |
1.428 |
0.025 |
| H12 |
2.239 |
-0.734 |
0.169 |
Atom - Atom Distances (Å)
| |
C1 |
C2 |
C3 |
O4 |
H5 |
H6 |
H7 |
H8 |
H9 |
H10 |
H11 |
H12 |
| C1 | | 1.5242 | 2.5256 | 2.9160 | 1.0894 | 1.0916 | 1.0886 | 2.1595 | 2.1625 | 2.7506 | 3.4708 | 3.7696 |
C2 | 1.5242 | | 1.5160 | 2.3761 | 2.1710 | 2.1659 | 2.1609 | 1.0921 | 1.0923 | 2.1490 | 2.1533 | 3.2201 | C3 | 2.5256 | 1.5160 | | 1.4177 | 3.4744 | 2.7883 | 2.7751 | 2.1286 | 2.1289 | 1.0967 | 1.0948 | 1.9575 | O4 | 2.9160 | 2.3761 | 1.4177 | | 3.8945 | 3.3269 | 2.5979 | 2.5970 | 3.3115 | 2.0732 | 2.0722 | 0.9579 | H5 | 1.0894 | 2.1710 | 3.4744 | 3.8945 | | 1.7639 | 1.7662 | 2.5044 | 2.5113 | 3.7544 | 4.3029 | 4.7916 | H6 | 1.0916 | 2.1659 | 2.7883 | 3.3269 | 1.7639 | | 1.7637 | 3.0684 | 2.5127 | 2.5632 | 3.7701 | 4.0145 | H7 | 1.0886 | 2.1609 | 2.7751 | 2.5979 | 1.7662 | 1.7637 | | 2.5103 | 3.0661 | 3.0917 | 3.7892 | 3.4288 | H8 | 2.1595 | 1.0921 | 2.1286 | 2.5970 | 2.5044 | 3.0684 | 2.5103 | | 1.7579 | 3.0407 | 2.4730 | 3.4883 | H9 | 2.1625 | 1.0923 | 2.1289 | 3.3115 | 2.5113 | 2.5127 | 3.0661 | 1.7579 | | 2.4871 | 2.4252 | 4.0496 | H10 | 2.7506 | 2.1490 | 1.0967 | 2.0732 | 3.7544 | 2.5632 | 3.0917 | 3.0407 | 2.4871 | | 1.7742 | 2.3172 | H11 | 3.4708 | 2.1533 | 1.0948 | 2.0722 | 4.3029 | 3.7701 | 3.7892 | 2.4730 | 2.4252 | 1.7742 | | 2.3427 | H12 | 3.7696 | 3.2201 | 1.9575 | 0.9579 | 4.7916 | 4.0145 | 3.4288 | 3.4883 | 4.0496 | 2.3172 | 2.3427 | |
 More geometry information
More geometry information
Calculated Bond Angles
| atom1 |
atom2 |
atom3 |
angle |
|
atom1 |
atom2 |
atom3 |
angle |
| C1 |
C2 |
C3 |
112.345 |
|
C1 |
C2 |
H8 |
110.158 |
| C1 |
C2 |
H9 |
110.388 |
|
C2 |
C1 |
H5 |
111.239 |
| C2 |
C1 |
H6 |
110.701 |
|
C2 |
C1 |
H7 |
110.482 |
| C2 |
C3 |
O4 |
108.125 |
|
C2 |
C3 |
H10 |
109.634 |
| C2 |
C3 |
H11 |
110.079 |
|
C3 |
C2 |
H8 |
108.303 |
| C3 |
C2 |
H9 |
108.310 |
|
C3 |
O4 |
H12 |
109.448 |
| O4 |
C3 |
H10 |
110.431 |
|
O4 |
C3 |
H11 |
110.461 |
| H5 |
C1 |
H6 |
107.944 |
|
H5 |
C1 |
H7 |
108.373 |
| H6 |
C1 |
H7 |
107.990 |
|
H8 |
C2 |
H9 |
107.169 |
| H10 |
C3 |
H11 |
108.111 |
|
Electronic energy levels
Charges, Dipole, Quadrupole and Polarizability
Charges from optimized geometry at M06-2X/6-311+G(3df,2p)
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.654 |
|
|
|
| 2 |
C |
-0.305 |
|
|
|
| 3 |
C |
-0.058 |
|
|
|
| 4 |
O |
-0.614 |
|
|
|
| 5 |
H |
0.183 |
|
|
|
| 6 |
H |
0.199 |
|
|
|
| 7 |
H |
0.218 |
|
|
|
| 8 |
H |
0.220 |
|
|
|
| 9 |
H |
0.199 |
|
|
|
| 10 |
H |
0.198 |
|
|
|
| 11 |
H |
0.180 |
|
|
|
| 12 |
H |
0.233 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.659 |
0.951 |
0.987 |
1.520 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-23.129 |
-0.171 |
2.016 |
| y |
-0.171 |
-27.435 |
-0.592 |
| z |
2.016 |
-0.592 |
-27.096 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
4.137 |
-0.171 |
2.016 |
| y |
-0.171 |
-2.323 |
-0.592 |
| z |
2.016 |
-0.592 |
-1.814 |
|
| Polar |
|---|
| 3z2-r2 | -3.628 |
|---|
| x2-y2 | 4.307 |
|---|
| xy | -0.171 |
|---|
| xz | 2.016 |
|---|
| yz | -0.592 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
7.070 |
0.080 |
0.083 |
| y |
0.080 |
6.354 |
-0.024 |
| z |
0.083 |
-0.024 |
5.944 |
<r2> (average value of r
2) Å
2
| <r2> |
94.456 |
| (<r2>)1/2 |
9.719 |
