Jump to
S1C2
Vibrational Frequencies calculated at B2PLYP=FULLultrafine/6-311+G(3df,2p)
Geometric Data calculated at B2PLYP=FULLultrafine/6-311+G(3df,2p)
Electronic energy levels
Charges, Dipole, Quadrupole and Polarizability
Jump to
S1C1
Energy calculated at B2PLYP=FULLultrafine/6-311+G(3df,2p)
| | hartrees |
|---|
| Energy at 0K | -194.279480 |
| Energy at 298.15K | |
| HF Energy | -194.004441 |
| Nuclear repulsion energy | 132.890930 |
The energy at 298.15K was derived from the energy at 0K
and an integrated heat capacity that used the calculated vibrational frequencies.
Vibrational Frequencies calculated at B2PLYP=FULLultrafine/6-311+G(3df,2p)
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3876 |
3876 |
34.74 |
104.26 |
0.23 |
0.37 |
| 2 |
A |
3139 |
3139 |
22.35 |
33.93 |
0.74 |
0.85 |
| 3 |
A |
3116 |
3116 |
44.20 |
55.81 |
0.55 |
0.71 |
| 4 |
A |
3085 |
3085 |
20.82 |
74.25 |
0.71 |
0.83 |
| 5 |
A |
3052 |
3052 |
53.19 |
159.72 |
0.06 |
0.11 |
| 6 |
A |
3050 |
3050 |
35.11 |
124.19 |
0.04 |
0.08 |
| 7 |
A |
3036 |
3036 |
15.04 |
94.34 |
0.58 |
0.73 |
| 8 |
A |
3003 |
3003 |
57.25 |
115.01 |
0.09 |
0.16 |
| 9 |
A |
1535 |
1535 |
2.81 |
3.30 |
0.75 |
0.86 |
| 10 |
A |
1525 |
1525 |
7.15 |
2.20 |
0.75 |
0.86 |
| 11 |
A |
1512 |
1512 |
7.29 |
6.41 |
0.75 |
0.86 |
| 12 |
A |
1493 |
1493 |
2.73 |
6.70 |
0.73 |
0.84 |
| 13 |
A |
1456 |
1456 |
4.84 |
0.97 |
0.15 |
0.27 |
| 14 |
A |
1422 |
1422 |
2.79 |
0.13 |
0.75 |
0.86 |
| 15 |
A |
1390 |
1390 |
1.21 |
0.80 |
0.74 |
0.85 |
| 16 |
A |
1331 |
1331 |
14.84 |
4.90 |
0.71 |
0.83 |
| 17 |
A |
1281 |
1281 |
0.37 |
2.95 |
0.73 |
0.85 |
| 18 |
A |
1249 |
1249 |
36.81 |
1.84 |
0.35 |
0.52 |
| 19 |
A |
1170 |
1170 |
5.30 |
1.15 |
0.08 |
0.15 |
| 20 |
A |
1122 |
1122 |
10.65 |
3.70 |
0.48 |
0.65 |
| 21 |
A |
1075 |
1075 |
42.74 |
2.93 |
0.59 |
0.74 |
| 22 |
A |
989 |
989 |
50.63 |
3.44 |
0.69 |
0.81 |
| 23 |
A |
933 |
933 |
2.14 |
0.39 |
0.71 |
0.83 |
| 24 |
A |
873 |
873 |
1.40 |
11.78 |
0.09 |
0.16 |
| 25 |
A |
779 |
779 |
0.85 |
0.40 |
0.25 |
0.40 |
| 26 |
A |
479 |
479 |
8.27 |
0.23 |
0.48 |
0.65 |
| 27 |
A |
328 |
328 |
5.43 |
0.29 |
0.29 |
0.45 |
| 28 |
A |
245 |
245 |
104.89 |
1.49 |
0.74 |
0.85 |
| 29 |
A |
225 |
225 |
3.64 |
0.05 |
0.48 |
0.64 |
| 30 |
A |
142 |
142 |
9.56 |
0.14 |
0.73 |
0.85 |
Unscaled Zero Point Vibrational Energy (zpe) 23955.3 cm
-1
Scaled (by 1) Zero Point Vibrational Energy (zpe) 23955.3 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Geometric Data calculated at B2PLYP=FULLultrafine/6-311+G(3df,2p)
Point Group is C1
Cartesians (Å)
| Atom |
x (Å) |
y (Å) |
z (Å) |
|---|
| C1 |
-1.538 |
-0.514 |
0.128 |
| C2 |
-0.633 |
0.638 |
-0.292 |
| C3 |
0.761 |
0.544 |
0.296 |
| O4 |
1.385 |
-0.631 |
-0.216 |
| H5 |
-2.529 |
-0.415 |
-0.312 |
| H6 |
-1.657 |
-0.541 |
1.212 |
| H7 |
-1.122 |
-1.468 |
-0.186 |
| H8 |
-0.545 |
0.669 |
-1.379 |
| H9 |
-1.066 |
1.590 |
0.019 |
| H10 |
0.702 |
0.498 |
1.388 |
| H11 |
1.342 |
1.430 |
0.025 |
| H12 |
2.256 |
-0.719 |
0.173 |
Atom - Atom Distances (Å)
| |
C1 |
C2 |
C3 |
O4 |
H5 |
H6 |
H7 |
H8 |
H9 |
H10 |
H11 |
H12 |
| C1 | | 1.5246 | 2.5367 | 2.9455 | 1.0888 | 1.0905 | 1.0873 | 2.1587 | 2.1599 | 2.7627 | 3.4765 | 3.8002 |
C2 | 1.5246 | | 1.5154 | 2.3843 | 2.1690 | 2.1684 | 2.1647 | 1.0912 | 1.0919 | 2.1506 | 2.1508 | 3.2253 | C3 | 2.5367 | 1.5154 | | 1.4255 | 3.4803 | 2.8042 | 2.7978 | 2.1280 | 2.1237 | 1.0952 | 1.0937 | 1.9614 | O4 | 2.9455 | 2.3843 | 1.4255 | | 3.9209 | 3.3614 | 2.6429 | 2.6017 | 3.3161 | 2.0775 | 2.0760 | 0.9586 | H5 | 1.0888 | 2.1690 | 3.4803 | 3.9209 | | 1.7604 | 1.7624 | 2.4999 | 2.5039 | 3.7636 | 4.3011 | 4.8194 | H6 | 1.0905 | 2.1684 | 2.8042 | 3.3614 | 1.7604 | | 1.7609 | 3.0686 | 2.5134 | 2.5839 | 3.7799 | 4.0525 | H7 | 1.0873 | 2.1647 | 2.7978 | 2.6429 | 1.7624 | 1.7609 | | 2.5149 | 3.0661 | 3.1103 | 3.8100 | 3.4787 | H8 | 2.1587 | 1.0912 | 2.1280 | 2.6017 | 2.4999 | 3.0686 | 2.5149 | | 1.7533 | 3.0408 | 2.4722 | 3.4910 | H9 | 2.1599 | 1.0919 | 2.1237 | 3.3161 | 2.5039 | 2.5134 | 3.0661 | 1.7533 | | 2.4892 | 2.4130 | 4.0493 | H10 | 2.7627 | 2.1506 | 1.0952 | 2.0775 | 3.7636 | 2.5839 | 3.1103 | 3.0408 | 2.4892 | | 1.7713 | 2.3183 | H11 | 3.4765 | 2.1508 | 1.0937 | 2.0760 | 4.3011 | 3.7799 | 3.8100 | 2.4722 | 2.4130 | 1.7713 | | 2.3411 | H12 | 3.8002 | 3.2253 | 1.9614 | 0.9586 | 4.8194 | 4.0525 | 3.4787 | 3.4910 | 4.0493 | 2.3183 | 2.3411 | |
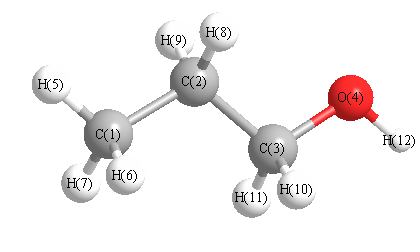 More geometry information
More geometry information
Calculated Bond Angles
| atom1 |
atom2 |
atom3 |
angle |
|
atom1 |
atom2 |
atom3 |
angle |
| C1 |
C2 |
C3 |
113.115 |
|
C1 |
C2 |
H8 |
110.121 |
| C1 |
C2 |
H9 |
110.179 |
|
C2 |
C1 |
H5 |
111.092 |
| C2 |
C1 |
H6 |
110.931 |
|
C2 |
C1 |
H7 |
110.839 |
| C2 |
C3 |
O4 |
108.295 |
|
C2 |
C3 |
H10 |
109.893 |
| C2 |
C3 |
H11 |
109.993 |
|
C3 |
C2 |
H8 |
108.357 |
| C3 |
C2 |
H9 |
107.980 |
|
C3 |
O4 |
H12 |
109.139 |
| O4 |
C3 |
H10 |
110.325 |
|
O4 |
C3 |
H11 |
110.296 |
| H5 |
C1 |
H6 |
107.757 |
|
H5 |
C1 |
H7 |
108.175 |
| H6 |
C1 |
H7 |
107.914 |
|
H8 |
C2 |
H9 |
106.867 |
| H10 |
C3 |
H11 |
108.040 |
|
Electronic energy levels
Charges, Dipole, Quadrupole and Polarizability
Charges from optimized geometry at B2PLYP=FULLultrafine/6-311+G(3df,2p)
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.474 |
|
|
|
| 2 |
C |
-0.186 |
|
|
|
| 3 |
C |
0.056 |
|
|
|
| 4 |
O |
-0.623 |
|
|
|
| 5 |
H |
0.138 |
|
|
|
| 6 |
H |
0.146 |
|
|
|
| 7 |
H |
0.162 |
|
|
|
| 8 |
H |
0.151 |
|
|
|
| 9 |
H |
0.139 |
|
|
|
| 10 |
H |
0.140 |
|
|
|
| 11 |
H |
0.129 |
|
|
|
| 12 |
H |
0.221 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.614 |
1.001 |
0.989 |
1.535 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-23.434 |
-0.083 |
2.045 |
| y |
-0.083 |
-27.678 |
-0.570 |
| z |
2.045 |
-0.570 |
-27.260 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
4.035 |
-0.083 |
2.045 |
| y |
-0.083 |
-2.331 |
-0.570 |
| z |
2.045 |
-0.570 |
-1.704 |
|
| Polar |
|---|
| 3z2-r2 | -3.408 |
|---|
| x2-y2 | 4.244 |
|---|
| xy | -0.083 |
|---|
| xz | 2.045 |
|---|
| yz | -0.570 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
7.141 |
0.066 |
0.064 |
| y |
0.066 |
6.453 |
-0.015 |
| z |
0.064 |
-0.015 |
6.043 |
<r2> (average value of r
2) Å
2
| <r2> |
95.317 |
| (<r2>)1/2 |
9.763 |
