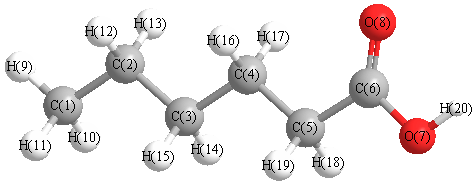
Vibrational Frequencies calculated at B3LYP/SDD
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3644 |
3503 |
32.27 |
|
|
|
| 2 |
A |
3124 |
3003 |
167.91 |
|
|
|
| 3 |
A |
3123 |
3002 |
59.25 |
|
|
|
| 4 |
A |
3118 |
2997 |
0.94 |
|
|
|
| 5 |
A |
3093 |
2973 |
2.09 |
|
|
|
| 6 |
A |
3078 |
2959 |
17.43 |
|
|
|
| 7 |
A |
3058 |
2940 |
45.83 |
|
|
|
| 8 |
A |
3051 |
2933 |
3.72 |
|
|
|
| 9 |
A |
3051 |
2933 |
3.74 |
|
|
|
| 10 |
A |
3031 |
2914 |
18.35 |
|
|
|
| 11 |
A |
3029 |
2912 |
70.20 |
|
|
|
| 12 |
A |
3014 |
2897 |
16.42 |
|
|
|
| 13 |
A |
1719 |
1653 |
236.69 |
|
|
|
| 14 |
A |
1535 |
1475 |
20.84 |
|
|
|
| 15 |
A |
1525 |
1466 |
3.43 |
|
|
|
| 16 |
A |
1521 |
1462 |
11.85 |
|
|
|
| 17 |
A |
1517 |
1458 |
1.31 |
|
|
|
| 18 |
A |
1513 |
1454 |
0.24 |
|
|
|
| 19 |
A |
1488 |
1430 |
26.41 |
|
|
|
| 20 |
A |
1436 |
1380 |
8.32 |
|
|
|
| 21 |
A |
1420 |
1366 |
34.82 |
|
|
|
| 22 |
A |
1403 |
1349 |
3.65 |
|
|
|
| 23 |
A |
1366 |
1313 |
29.52 |
|
|
|
| 24 |
A |
1333 |
1281 |
0.83 |
|
|
|
| 25 |
A |
1328 |
1276 |
0.00 |
|
|
|
| 26 |
A |
1307 |
1257 |
22.10 |
|
|
|
| 27 |
A |
1296 |
1246 |
0.07 |
|
|
|
| 28 |
A |
1273 |
1224 |
1.49 |
|
|
|
| 29 |
A |
1238 |
1190 |
0.01 |
|
|
|
| 30 |
A |
1155 |
1110 |
1.38 |
|
|
|
| 31 |
A |
1150 |
1105 |
45.90 |
|
|
|
| 32 |
A |
1102 |
1059 |
94.42 |
|
|
|
| 33 |
A |
1089 |
1047 |
13.39 |
|
|
|
| 34 |
A |
1070 |
1029 |
116.45 |
|
|
|
| 35 |
A |
1021 |
981 |
63.53 |
|
|
|
| 36 |
A |
987 |
949 |
0.11 |
|
|
|
| 37 |
A |
926 |
890 |
1.51 |
|
|
|
| 38 |
A |
867 |
834 |
6.99 |
|
|
|
| 39 |
A |
864 |
831 |
1.94 |
|
|
|
| 40 |
A |
776 |
746 |
1.21 |
|
|
|
| 41 |
A |
743 |
715 |
15.82 |
|
|
|
| 42 |
A |
644 |
619 |
142.90 |
|
|
|
| 43 |
A |
608 |
584 |
18.66 |
|
|
|
| 44 |
A |
516 |
496 |
33.94 |
|
|
|
| 45 |
A |
498 |
479 |
31.42 |
|
|
|
| 46 |
A |
402 |
387 |
1.23 |
|
|
|
| 47 |
A |
300 |
289 |
2.67 |
|
|
|
| 48 |
A |
229 |
220 |
0.01 |
|
|
|
| 49 |
A |
222 |
213 |
2.07 |
|
|
|
| 50 |
A |
129 |
124 |
0.02 |
|
|
|
| 51 |
A |
109 |
105 |
0.17 |
|
|
|
| 52 |
A |
97 |
93 |
0.99 |
|
|
|
| 53 |
A |
57 |
55 |
0.13 |
|
|
|
| 54 |
A |
34 |
33 |
0.28 |
|
|
|
Unscaled Zero Point Vibrational Energy (zpe) 38612.2 cm
-1
Scaled (by 0.9613) Zero Point Vibrational Energy (zpe) 37117.9 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Charges, Dipole, Quadrupole and Polarizability
Charges from optimized geometry at B3LYP/SDD
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.647 |
|
|
|
| 2 |
C |
-0.310 |
|
|
|
| 3 |
C |
-0.298 |
|
|
|
| 4 |
C |
-0.319 |
|
|
|
| 5 |
C |
-0.443 |
|
|
|
| 6 |
C |
0.218 |
|
|
|
| 7 |
O |
-0.452 |
|
|
|
| 8 |
O |
-0.261 |
|
|
|
| 9 |
H |
0.201 |
|
|
|
| 10 |
H |
0.193 |
|
|
|
| 11 |
H |
0.193 |
|
|
|
| 12 |
H |
0.178 |
|
|
|
| 13 |
H |
0.178 |
|
|
|
| 14 |
H |
0.166 |
|
|
|
| 15 |
H |
0.166 |
|
|
|
| 16 |
H |
0.194 |
|
|
|
| 17 |
H |
0.194 |
|
|
|
| 18 |
H |
0.234 |
|
|
|
| 19 |
H |
0.234 |
|
|
|
| 20 |
H |
0.383 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.560 |
-1.490 |
0.000 |
1.591 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-45.901 |
3.003 |
0.001 |
| y |
3.003 |
-56.086 |
0.001 |
| z |
0.001 |
0.001 |
-48.590 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
6.437 |
3.003 |
0.001 |
| y |
3.003 |
-8.840 |
0.001 |
| z |
0.001 |
0.001 |
2.403 |
|
| Polar |
|---|
| 3z2-r2 | 4.807 |
|---|
| x2-y2 | 10.185 |
|---|
| xy | 3.003 |
|---|
| xz | 0.001 |
|---|
| yz | 0.001 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
12.660 |
0.048 |
0.000 |
| y |
0.048 |
9.891 |
-0.000 |
| z |
0.000 |
-0.000 |
7.863 |
<r2> (average value of r
2) Å
2
| <r2> |
521.254 |
| (<r2>)1/2 |
22.831 |