Jump to
S1C2
Energy calculated at CCSD(T)/6-311G**
| | hartrees |
|---|
| Energy at 0K | -193.899549 |
| Energy at 298.15K | -193.908430 |
| Nuclear repulsion energy | 130.124985 |
The energy at 298.15K was derived from the energy at 0K
and an integrated heat capacity that used the calculated vibrational frequencies.
Vibrational Frequencies calculated at CCSD(T)/6-311G**
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A' |
3890 |
3753 |
|
|
|
|
| 2 |
A' |
3113 |
3004 |
|
|
|
|
| 3 |
A' |
3049 |
2941 |
|
|
|
|
| 4 |
A' |
3031 |
2924 |
|
|
|
|
| 5 |
A' |
2981 |
2876 |
|
|
|
|
| 6 |
A' |
1543 |
1489 |
|
|
|
|
| 7 |
A' |
1522 |
1468 |
|
|
|
|
| 8 |
A' |
1506 |
1452 |
|
|
|
|
| 9 |
A' |
1485 |
1433 |
|
|
|
|
| 10 |
A' |
1429 |
1379 |
|
|
|
|
| 11 |
A' |
1364 |
1315 |
|
|
|
|
| 12 |
A' |
1287 |
1241 |
|
|
|
|
| 13 |
A' |
1116 |
1076 |
|
|
|
|
| 14 |
A' |
1093 |
1054 |
|
|
|
|
| 15 |
A' |
1058 |
1021 |
|
|
|
|
| 16 |
A' |
899 |
867 |
|
|
|
|
| 17 |
A' |
459 |
443 |
|
|
|
|
| 18 |
A' |
271 |
262 |
|
|
|
|
| 19 |
A" |
3109 |
2999 |
|
|
|
|
| 20 |
A" |
3085 |
2976 |
|
|
|
|
| 21 |
A" |
3014 |
2908 |
|
|
|
|
| 22 |
A" |
1511 |
1457 |
|
|
|
|
| 23 |
A" |
1328 |
1282 |
|
|
|
|
| 24 |
A" |
1282 |
1237 |
|
|
|
|
| 25 |
A" |
1202 |
1160 |
|
|
|
|
| 26 |
A" |
909 |
877 |
|
|
|
|
| 27 |
A" |
776 |
749 |
|
|
|
|
| 28 |
A" |
243 |
235 |
|
|
|
|
| 29 |
A" |
229 |
220 |
|
|
|
|
| 30 |
A" |
121 |
117 |
|
|
|
|
Unscaled Zero Point Vibrational Energy (zpe) 23951.5 cm
-1
Scaled (by 0.9647) Zero Point Vibrational Energy (zpe) 23106.0 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Geometric Data calculated at CCSD(T)/6-311G**
Point Group is Cs
Cartesians (Å)
| Atom |
x (Å) |
y (Å) |
z (Å) |
|---|
| C1 |
-1.456 |
1.226 |
0.000 |
| C2 |
0.000 |
0.743 |
0.000 |
| C3 |
0.101 |
-0.777 |
0.000 |
| O4 |
1.483 |
-1.126 |
0.000 |
| H5 |
-1.508 |
2.321 |
0.000 |
| H6 |
-1.991 |
0.862 |
0.888 |
| H7 |
-1.991 |
0.862 |
-0.888 |
| H8 |
0.529 |
1.124 |
0.885 |
| H9 |
0.529 |
1.124 |
-0.885 |
| H10 |
-0.411 |
-1.178 |
0.892 |
| H11 |
-0.411 |
-1.178 |
-0.892 |
| H12 |
1.525 |
-2.084 |
0.000 |
Atom - Atom Distances (Å)
| |
C1 |
C2 |
C3 |
O4 |
H5 |
H6 |
H7 |
H8 |
H9 |
H10 |
H11 |
H12 |
| C1 | | 1.5341 | 2.5370 | 3.7638 | 1.0968 | 1.0986 | 1.0986 | 2.1757 | 2.1757 | 2.7688 | 2.7688 | 4.4546 |
C2 | 1.5341 | | 1.5239 | 2.3857 | 2.1827 | 2.1833 | 2.1833 | 1.0985 | 1.0985 | 2.1578 | 2.1578 | 3.2126 | C3 | 2.5370 | 1.5239 | | 1.4250 | 3.4914 | 2.8023 | 2.8023 | 2.1398 | 2.1398 | 1.1040 | 1.1040 | 1.9330 | O4 | 3.7638 | 2.3857 | 1.4250 | | 4.5636 | 4.0997 | 4.0997 | 2.5984 | 2.5984 | 2.0940 | 2.0940 | 0.9594 | H5 | 1.0968 | 2.1827 | 3.4914 | 4.5636 | | 1.7749 | 1.7749 | 2.5232 | 2.5232 | 3.7741 | 3.7741 | 5.3487 | H6 | 1.0986 | 2.1833 | 2.8023 | 4.0997 | 1.7749 | | 1.7765 | 2.5334 | 3.0920 | 2.5805 | 3.1352 | 4.6726 | H7 | 1.0986 | 2.1833 | 2.8023 | 4.0997 | 1.7749 | 1.7765 | | 3.0920 | 2.5334 | 3.1352 | 2.5805 | 4.6726 | H8 | 2.1757 | 1.0985 | 2.1398 | 2.5984 | 2.5232 | 2.5334 | 3.0920 | | 1.7690 | 2.4860 | 3.0557 | 3.4734 | H9 | 2.1757 | 1.0985 | 2.1398 | 2.5984 | 2.5232 | 3.0920 | 2.5334 | 1.7690 | | 3.0557 | 2.4860 | 3.4734 | H10 | 2.7688 | 2.1578 | 1.1040 | 2.0940 | 3.7741 | 2.5805 | 3.1352 | 2.4860 | 3.0557 | | 1.7847 | 2.3164 | H11 | 2.7688 | 2.1578 | 1.1040 | 2.0940 | 3.7741 | 3.1352 | 2.5805 | 3.0557 | 2.4860 | 1.7847 | | 2.3164 | H12 | 4.4546 | 3.2126 | 1.9330 | 0.9594 | 5.3487 | 4.6726 | 4.6726 | 3.4734 | 3.4734 | 2.3164 | 2.3164 | |
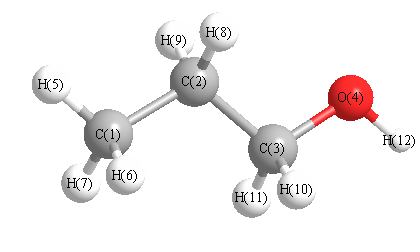 More geometry information
More geometry information
Calculated Bond Angles
| atom1 |
atom2 |
atom3 |
angle |
|
atom1 |
atom2 |
atom3 |
angle |
| C1 |
C2 |
C3 |
112.123 |
|
C1 |
C2 |
H8 |
110.379 |
| C1 |
C2 |
H9 |
110.379 |
|
C2 |
C1 |
H5 |
111.039 |
| C2 |
C1 |
H6 |
110.979 |
|
C2 |
C1 |
H7 |
110.979 |
| C2 |
C3 |
O4 |
107.952 |
|
C2 |
C3 |
H10 |
109.354 |
| C2 |
C3 |
H11 |
109.354 |
|
C3 |
C2 |
H8 |
108.272 |
| C3 |
C2 |
H9 |
108.272 |
|
C3 |
O4 |
H12 |
106.687 |
| O4 |
C3 |
H10 |
111.149 |
|
O4 |
C3 |
H11 |
111.149 |
| H5 |
C1 |
H6 |
107.896 |
|
H5 |
C1 |
H7 |
107.896 |
| H6 |
C1 |
H7 |
107.911 |
|
H8 |
C2 |
H9 |
107.257 |
| H10 |
C3 |
H11 |
107.862 |
|
Electronic energy levels
Charges, Dipole, Quadrupole and Polarizability
Jump to
S1C1
Energy calculated at CCSD(T)/6-311G**
| | hartrees |
|---|
| Energy at 0K | -193.900234 |
| Energy at 298.15K | |
| HF Energy | -193.168143 |
| Nuclear repulsion energy | 132.530002 |
The energy at 298.15K was derived from the energy at 0K
and an integrated heat capacity that used the calculated vibrational frequencies.
Vibrational Frequencies calculated at CCSD(T)/6-311G**
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3892 |
3754 |
|
|
|
|
| 2 |
A |
3134 |
3023 |
|
|
|
|
| 3 |
A |
3106 |
2996 |
|
|
|
|
| 4 |
A |
3081 |
2972 |
|
|
|
|
| 5 |
A |
3042 |
2934 |
|
|
|
|
| 6 |
A |
3035 |
2927 |
|
|
|
|
| 7 |
A |
3020 |
2914 |
|
|
|
|
| 8 |
A |
2984 |
2879 |
|
|
|
|
| 9 |
A |
1539 |
1484 |
|
|
|
|
| 10 |
A |
1521 |
1468 |
|
|
|
|
| 11 |
A |
1506 |
1453 |
|
|
|
|
| 12 |
A |
1489 |
1437 |
|
|
|
|
| 13 |
A |
1478 |
1426 |
|
|
|
|
| 14 |
A |
1429 |
1379 |
|
|
|
|
| 15 |
A |
1389 |
1340 |
|
|
|
|
| 16 |
A |
1339 |
1291 |
|
|
|
|
| 17 |
A |
1288 |
1242 |
|
|
|
|
| 18 |
A |
1272 |
1227 |
|
|
|
|
| 19 |
A |
1177 |
1136 |
|
|
|
|
| 20 |
A |
1139 |
1099 |
|
|
|
|
| 21 |
A |
1091 |
1052 |
|
|
|
|
| 22 |
A |
1000 |
965 |
|
|
|
|
| 23 |
A |
934 |
901 |
|
|
|
|
| 24 |
A |
882 |
850 |
|
|
|
|
| 25 |
A |
780 |
753 |
|
|
|
|
| 26 |
A |
481 |
464 |
|
|
|
|
| 27 |
A |
330 |
318 |
|
|
|
|
| 28 |
A |
238 |
230 |
|
|
|
|
| 29 |
A |
225 |
217 |
|
|
|
|
| 30 |
A |
141 |
136 |
|
|
|
|
Unscaled Zero Point Vibrational Energy (zpe) 23979.1 cm
-1
Scaled (by 0.9647) Zero Point Vibrational Energy (zpe) 23132.6 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Geometric Data calculated at CCSD(T)/6-311G**
Point Group is C1
Cartesians (Å)
| Atom |
x (Å) |
y (Å) |
z (Å) |
|---|
| C1 |
-1.532 |
-0.520 |
0.130 |
| C2 |
-0.634 |
0.650 |
-0.292 |
| C3 |
0.771 |
0.546 |
0.294 |
| O4 |
1.370 |
-0.642 |
-0.220 |
| H5 |
-2.532 |
-0.430 |
-0.312 |
| H6 |
-1.643 |
-0.545 |
1.223 |
| H7 |
-1.091 |
-1.471 |
-0.189 |
| H8 |
-0.547 |
0.686 |
-1.387 |
| H9 |
-1.074 |
1.604 |
0.034 |
| H10 |
0.705 |
0.510 |
1.396 |
| H11 |
1.358 |
1.437 |
0.014 |
| H12 |
2.234 |
-0.716 |
0.189 |
Atom - Atom Distances (Å)
| |
C1 |
C2 |
C3 |
O4 |
H5 |
H6 |
H7 |
H8 |
H9 |
H10 |
H11 |
H12 |
| C1 | | 1.5340 | 2.5423 | 2.9250 | 1.0973 | 1.0991 | 1.0952 | 2.1731 | 2.1748 | 2.7690 | 3.4914 | 3.7714 |
C2 | 1.5340 | | 1.5251 | 2.3851 | 2.1842 | 2.1772 | 2.1723 | 1.0991 | 1.1000 | 2.1588 | 2.1628 | 3.2130 | C3 | 2.5423 | 1.5251 | | 1.4265 | 3.4967 | 2.8069 | 2.7870 | 2.1405 | 2.1425 | 1.1042 | 1.1027 | 1.9357 | O4 | 2.9250 | 2.3851 | 1.4265 | | 3.9085 | 3.3423 | 2.5971 | 2.6076 | 3.3291 | 2.0926 | 2.0915 | 0.9593 | H5 | 1.0973 | 2.1842 | 3.4967 | 3.9085 | | 1.7774 | 1.7815 | 2.5177 | 2.5263 | 3.7790 | 4.3267 | 4.8010 | H6 | 1.0991 | 2.1772 | 2.8069 | 3.3423 | 1.7774 | | 1.7766 | 3.0863 | 2.5205 | 2.5802 | 3.7937 | 4.0167 | H7 | 1.0952 | 2.1723 | 2.7870 | 2.5971 | 1.7815 | 1.7766 | | 2.5262 | 3.0830 | 3.1083 | 3.8067 | 3.4309 | H8 | 2.1731 | 1.0991 | 2.1405 | 2.6076 | 2.5177 | 3.0863 | 2.5262 | | 1.7719 | 3.0566 | 2.4810 | 3.4909 | H9 | 2.1748 | 1.1000 | 2.1425 | 3.3291 | 2.5263 | 2.5205 | 3.0830 | 1.7719 | | 2.4938 | 2.4380 | 4.0440 | H10 | 2.7690 | 2.1588 | 1.1042 | 2.0926 | 3.7790 | 2.5802 | 3.1083 | 3.0566 | 2.4938 | | 1.7870 | 2.3014 | H11 | 3.4914 | 2.1628 | 1.1027 | 2.0915 | 4.3267 | 3.7937 | 3.8067 | 2.4810 | 2.4380 | 1.7870 | | 2.3308 | H12 | 3.7714 | 3.2130 | 1.9357 | 0.9593 | 4.8010 | 4.0167 | 3.4309 | 3.4909 | 4.0440 | 2.3014 | 2.3308 | |
 More geometry information
More geometry information
Calculated Bond Angles
| atom1 |
atom2 |
atom3 |
angle |
|
atom1 |
atom2 |
atom3 |
angle |
| C1 |
C2 |
C3 |
112.418 |
|
C1 |
C2 |
H8 |
110.143 |
| C1 |
C2 |
H9 |
110.221 |
|
C2 |
C1 |
H5 |
111.133 |
| C2 |
C1 |
H6 |
110.465 |
|
C2 |
C1 |
H7 |
110.306 |
| C2 |
C3 |
O4 |
107.771 |
|
C2 |
C3 |
H10 |
109.337 |
| C2 |
C3 |
H11 |
109.741 |
|
C3 |
C2 |
H8 |
108.214 |
| C3 |
C2 |
H9 |
108.318 |
|
C3 |
O4 |
H12 |
106.816 |
| O4 |
C3 |
H10 |
110.917 |
|
O4 |
C3 |
H11 |
110.921 |
| H5 |
C1 |
H6 |
108.042 |
|
H5 |
C1 |
H7 |
108.690 |
| H6 |
C1 |
H7 |
108.115 |
|
H8 |
C2 |
H9 |
107.366 |
| H10 |
C3 |
H11 |
108.143 |
|
Electronic energy levels
Charges, Dipole, Quadrupole and Polarizability
