Vibrational Frequencies calculated at BLYP/6-311G*
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3559 |
3550 |
12.58 |
|
|
|
| 2 |
A |
3009 |
3001 |
54.77 |
|
|
|
| 3 |
A |
3005 |
2997 |
124.61 |
|
|
|
| 4 |
A |
2998 |
2990 |
22.75 |
|
|
|
| 5 |
A |
2971 |
2964 |
1.07 |
|
|
|
| 6 |
A |
2964 |
2956 |
56.27 |
|
|
|
| 7 |
A |
2961 |
2954 |
19.93 |
|
|
|
| 8 |
A |
2952 |
2945 |
2.34 |
|
|
|
| 9 |
A |
2949 |
2942 |
46.66 |
|
|
|
| 10 |
A |
2936 |
2929 |
4.52 |
|
|
|
| 11 |
A |
2936 |
2928 |
34.40 |
|
|
|
| 12 |
A |
2920 |
2912 |
14.04 |
|
|
|
| 13 |
A |
1746 |
1741 |
233.98 |
|
|
|
| 14 |
A |
1493 |
1489 |
8.63 |
|
|
|
| 15 |
A |
1483 |
1479 |
8.05 |
|
|
|
| 16 |
A |
1482 |
1479 |
2.83 |
|
|
|
| 17 |
A |
1470 |
1466 |
0.72 |
|
|
|
| 18 |
A |
1464 |
1460 |
0.30 |
|
|
|
| 19 |
A |
1441 |
1437 |
14.85 |
|
|
|
| 20 |
A |
1390 |
1387 |
1.32 |
|
|
|
| 21 |
A |
1375 |
1371 |
19.24 |
|
|
|
| 22 |
A |
1365 |
1361 |
1.72 |
|
|
|
| 23 |
A |
1329 |
1326 |
22.37 |
|
|
|
| 24 |
A |
1318 |
1314 |
0.74 |
|
|
|
| 25 |
A |
1313 |
1309 |
0.09 |
|
|
|
| 26 |
A |
1287 |
1284 |
0.49 |
|
|
|
| 27 |
A |
1274 |
1270 |
0.03 |
|
|
|
| 28 |
A |
1244 |
1241 |
12.02 |
|
|
|
| 29 |
A |
1203 |
1200 |
0.05 |
|
|
|
| 30 |
A |
1117 |
1114 |
0.63 |
|
|
|
| 31 |
A |
1113 |
1111 |
105.50 |
|
|
|
| 32 |
A |
1076 |
1074 |
170.98 |
|
|
|
| 33 |
A |
1028 |
1025 |
0.70 |
|
|
|
| 34 |
A |
1020 |
1017 |
26.92 |
|
|
|
| 35 |
A |
978 |
976 |
21.26 |
|
|
|
| 36 |
A |
962 |
959 |
0.22 |
|
|
|
| 37 |
A |
889 |
887 |
2.26 |
|
|
|
| 38 |
A |
842 |
840 |
5.04 |
|
|
|
| 39 |
A |
832 |
829 |
2.56 |
|
|
|
| 40 |
A |
751 |
749 |
3.76 |
|
|
|
| 41 |
A |
717 |
715 |
12.26 |
|
|
|
| 42 |
A |
668 |
667 |
90.00 |
|
|
|
| 43 |
A |
612 |
610 |
21.49 |
|
|
|
| 44 |
A |
513 |
511 |
19.67 |
|
|
|
| 45 |
A |
492 |
491 |
22.37 |
|
|
|
| 46 |
A |
395 |
394 |
0.80 |
|
|
|
| 47 |
A |
294 |
293 |
1.64 |
|
|
|
| 48 |
A |
238 |
237 |
0.00 |
|
|
|
| 49 |
A |
221 |
220 |
1.82 |
|
|
|
| 50 |
A |
136 |
136 |
0.02 |
|
|
|
| 51 |
A |
114 |
114 |
0.18 |
|
|
|
| 52 |
A |
96 |
96 |
0.82 |
|
|
|
| 53 |
A |
59 |
59 |
0.04 |
|
|
|
| 54 |
A |
36 |
36 |
0.21 |
|
|
|
Unscaled Zero Point Vibrational Energy (zpe) 37514.6 cm
-1
Scaled (by 0.9975) Zero Point Vibrational Energy (zpe) 37420.8 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Charges, Dipole, Quadrupole and Polarizability
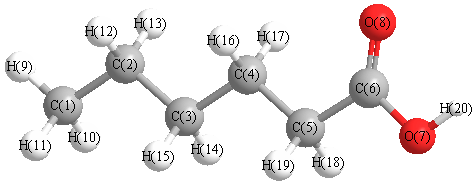
Charges from optimized geometry at BLYP/6-311G*
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.584 |
|
|
|
| 2 |
C |
-0.367 |
|
|
|
| 3 |
C |
-0.369 |
|
|
|
| 4 |
C |
-0.371 |
|
|
|
| 5 |
C |
-0.474 |
|
|
|
| 6 |
C |
0.385 |
|
|
|
| 7 |
O |
-0.470 |
|
|
|
| 8 |
O |
-0.295 |
|
|
|
| 9 |
H |
0.198 |
|
|
|
| 10 |
H |
0.192 |
|
|
|
| 11 |
H |
0.192 |
|
|
|
| 12 |
H |
0.189 |
|
|
|
| 13 |
H |
0.189 |
|
|
|
| 14 |
H |
0.184 |
|
|
|
| 15 |
H |
0.184 |
|
|
|
| 16 |
H |
0.201 |
|
|
|
| 17 |
H |
0.201 |
|
|
|
| 18 |
H |
0.224 |
|
|
|
| 19 |
H |
0.224 |
|
|
|
| 20 |
H |
0.368 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.212 |
-1.244 |
0.000 |
1.262 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-46.960 |
2.569 |
-0.001 |
| y |
2.569 |
-56.131 |
0.001 |
| z |
-0.001 |
0.001 |
-49.825 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
6.018 |
2.569 |
-0.001 |
| y |
2.569 |
-7.739 |
0.001 |
| z |
-0.001 |
0.001 |
1.721 |
|
| Polar |
|---|
| 3z2-r2 | 3.443 |
|---|
| x2-y2 | 9.171 |
|---|
| xy | 2.569 |
|---|
| xz | -0.001 |
|---|
| yz | 0.001 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
14.577 |
0.083 |
0.000 |
| y |
0.083 |
10.911 |
-0.000 |
| z |
0.000 |
-0.000 |
8.907 |
<r2> (average value of r
2) Å
2
| <r2> |
522.168 |
| (<r2>)1/2 |
22.851 |