Vibrational Frequencies calculated at mPW1PW91/6-31G
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3693 |
3496 |
0.48 |
|
|
|
| 2 |
A |
3570 |
3379 |
0.26 |
|
|
|
| 3 |
A |
3129 |
2962 |
46.28 |
|
|
|
| 4 |
A |
3115 |
2949 |
79.63 |
|
|
|
| 5 |
A |
3107 |
2941 |
38.21 |
|
|
|
| 6 |
A |
3106 |
2940 |
55.08 |
|
|
|
| 7 |
A |
3096 |
2931 |
64.41 |
|
|
|
| 8 |
A |
3053 |
2890 |
80.05 |
|
|
|
| 9 |
A |
3050 |
2887 |
14.53 |
|
|
|
| 10 |
A |
3048 |
2885 |
25.63 |
|
|
|
| 11 |
A |
3047 |
2884 |
12.67 |
|
|
|
| 12 |
A |
3034 |
2872 |
18.55 |
|
|
|
| 13 |
A |
3030 |
2869 |
11.35 |
|
|
|
| 14 |
A |
1715 |
1623 |
38.72 |
|
|
|
| 15 |
A |
1552 |
1469 |
3.00 |
|
|
|
| 16 |
A |
1545 |
1462 |
17.19 |
|
|
|
| 17 |
A |
1537 |
1455 |
5.61 |
|
|
|
| 18 |
A |
1535 |
1453 |
2.34 |
|
|
|
| 19 |
A |
1530 |
1448 |
2.05 |
|
|
|
| 20 |
A |
1446 |
1369 |
5.55 |
|
|
|
| 21 |
A |
1413 |
1337 |
0.65 |
|
|
|
| 22 |
A |
1412 |
1337 |
1.45 |
|
|
|
| 23 |
A |
1404 |
1329 |
0.95 |
|
|
|
| 24 |
A |
1398 |
1323 |
1.08 |
|
|
|
| 25 |
A |
1395 |
1320 |
1.82 |
|
|
|
| 26 |
A |
1375 |
1302 |
0.25 |
|
|
|
| 27 |
A |
1332 |
1261 |
1.38 |
|
|
|
| 28 |
A |
1319 |
1249 |
2.18 |
|
|
|
| 29 |
A |
1309 |
1239 |
2.18 |
|
|
|
| 30 |
A |
1256 |
1189 |
2.01 |
|
|
|
| 31 |
A |
1227 |
1162 |
1.83 |
|
|
|
| 32 |
A |
1168 |
1106 |
17.16 |
|
|
|
| 33 |
A |
1142 |
1081 |
0.75 |
|
|
|
| 34 |
A |
1130 |
1070 |
3.83 |
|
|
|
| 35 |
A |
1105 |
1046 |
0.70 |
|
|
|
| 36 |
A |
1077 |
1020 |
1.92 |
|
|
|
| 37 |
A |
1068 |
1011 |
4.68 |
|
|
|
| 38 |
A |
1018 |
964 |
1.95 |
|
|
|
| 39 |
A |
956 |
905 |
0.44 |
|
|
|
| 40 |
A |
926 |
877 |
14.20 |
|
|
|
| 41 |
A |
905 |
857 |
1.75 |
|
|
|
| 42 |
A |
875 |
828 |
1.75 |
|
|
|
| 43 |
A |
817 |
773 |
1.67 |
|
|
|
| 44 |
A |
805 |
762 |
1.83 |
|
|
|
| 45 |
A |
631 |
597 |
283.06 |
|
|
|
| 46 |
A |
563 |
533 |
5.22 |
|
|
|
| 47 |
A |
466 |
441 |
2.29 |
|
|
|
| 48 |
A |
459 |
434 |
7.11 |
|
|
|
| 49 |
A |
414 |
392 |
0.10 |
|
|
|
| 50 |
A |
347 |
328 |
16.77 |
|
|
|
| 51 |
A |
332 |
314 |
2.20 |
|
|
|
| 52 |
A |
247 |
234 |
23.42 |
|
|
|
| 53 |
A |
219 |
208 |
25.71 |
|
|
|
| 54 |
A |
158 |
150 |
0.26 |
|
|
|
Unscaled Zero Point Vibrational Energy (zpe) 41802.3 cm
-1
Scaled (by 0.9466) Zero Point Vibrational Energy (zpe) 39570.1 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Charges, Dipole, Quadrupole and Polarizability
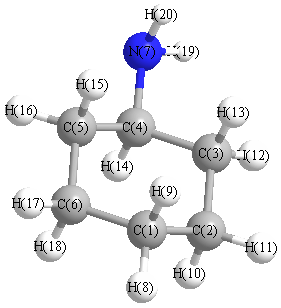
Charges from optimized geometry at mPW1PW91/6-31G
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.309 |
|
|
|
| 2 |
C |
-0.315 |
|
|
|
| 3 |
C |
-0.288 |
|
|
|
| 4 |
C |
-0.027 |
|
|
|
| 5 |
C |
-0.298 |
|
|
|
| 6 |
C |
-0.313 |
|
|
|
| 7 |
N |
-0.736 |
|
|
|
| 8 |
H |
0.157 |
|
|
|
| 9 |
H |
0.153 |
|
|
|
| 10 |
H |
0.155 |
|
|
|
| 11 |
H |
0.158 |
|
|
|
| 12 |
H |
0.175 |
|
|
|
| 13 |
H |
0.141 |
|
|
|
| 14 |
H |
0.160 |
|
|
|
| 15 |
H |
0.142 |
|
|
|
| 16 |
H |
0.147 |
|
|
|
| 17 |
H |
0.157 |
|
|
|
| 18 |
H |
0.153 |
|
|
|
| 19 |
H |
0.297 |
|
|
|
| 20 |
H |
0.293 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.298 |
1.046 |
0.501 |
1.198 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-45.946 |
-3.893 |
-1.736 |
| y |
-3.893 |
-45.955 |
-0.919 |
| z |
-1.736 |
-0.919 |
-43.857 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
-1.041 |
-3.893 |
-1.736 |
| y |
-3.893 |
-1.054 |
-0.919 |
| z |
-1.736 |
-0.919 |
2.094 |
|
| Polar |
|---|
| 3z2-r2 | 4.188 |
|---|
| x2-y2 | 0.009 |
|---|
| xy | -3.893 |
|---|
| xz | -1.736 |
|---|
| yz | -0.919 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
10.849 |
-0.173 |
-0.111 |
| y |
-0.173 |
9.759 |
-0.186 |
| z |
-0.111 |
-0.186 |
9.070 |
<r2> (average value of r
2) Å
2
| <r2> |
233.238 |
| (<r2>)1/2 |
15.272 |