Vibrational Frequencies calculated at B1B95/6-31G(2df,p)
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3600 |
3447 |
0.09 |
|
|
|
| 2 |
A |
3515 |
3367 |
0.78 |
|
|
|
| 3 |
A |
3114 |
2982 |
39.24 |
|
|
|
| 4 |
A |
3106 |
2975 |
53.54 |
|
|
|
| 5 |
A |
3101 |
2970 |
37.57 |
|
|
|
| 6 |
A |
3100 |
2969 |
47.96 |
|
|
|
| 7 |
A |
3084 |
2954 |
54.23 |
|
|
|
| 8 |
A |
3047 |
2919 |
47.89 |
|
|
|
| 9 |
A |
3045 |
2916 |
13.83 |
|
|
|
| 10 |
A |
3041 |
2912 |
19.19 |
|
|
|
| 11 |
A |
3034 |
2906 |
53.55 |
|
|
|
| 12 |
A |
3025 |
2897 |
15.48 |
|
|
|
| 13 |
A |
3017 |
2890 |
1.87 |
|
|
|
| 14 |
A |
1657 |
1587 |
26.09 |
|
|
|
| 15 |
A |
1499 |
1436 |
1.64 |
|
|
|
| 16 |
A |
1484 |
1422 |
11.72 |
|
|
|
| 17 |
A |
1481 |
1418 |
3.21 |
|
|
|
| 18 |
A |
1479 |
1417 |
0.80 |
|
|
|
| 19 |
A |
1472 |
1410 |
0.12 |
|
|
|
| 20 |
A |
1422 |
1362 |
4.60 |
|
|
|
| 21 |
A |
1384 |
1325 |
3.77 |
|
|
|
| 22 |
A |
1381 |
1323 |
0.69 |
|
|
|
| 23 |
A |
1379 |
1321 |
3.06 |
|
|
|
| 24 |
A |
1366 |
1309 |
0.49 |
|
|
|
| 25 |
A |
1358 |
1300 |
3.11 |
|
|
|
| 26 |
A |
1326 |
1270 |
0.83 |
|
|
|
| 27 |
A |
1298 |
1243 |
0.77 |
|
|
|
| 28 |
A |
1284 |
1230 |
2.60 |
|
|
|
| 29 |
A |
1281 |
1227 |
2.23 |
|
|
|
| 30 |
A |
1225 |
1173 |
5.77 |
|
|
|
| 31 |
A |
1199 |
1149 |
1.76 |
|
|
|
| 32 |
A |
1149 |
1100 |
9.11 |
|
|
|
| 33 |
A |
1131 |
1083 |
4.91 |
|
|
|
| 34 |
A |
1096 |
1050 |
1.50 |
|
|
|
| 35 |
A |
1067 |
1021 |
0.31 |
|
|
|
| 36 |
A |
1055 |
1010 |
4.12 |
|
|
|
| 37 |
A |
1046 |
1002 |
0.17 |
|
|
|
| 38 |
A |
991 |
950 |
1.01 |
|
|
|
| 39 |
A |
945 |
905 |
34.22 |
|
|
|
| 40 |
A |
908 |
869 |
60.04 |
|
|
|
| 41 |
A |
895 |
857 |
19.10 |
|
|
|
| 42 |
A |
874 |
837 |
56.73 |
|
|
|
| 43 |
A |
860 |
824 |
0.85 |
|
|
|
| 44 |
A |
795 |
761 |
3.33 |
|
|
|
| 45 |
A |
786 |
753 |
0.14 |
|
|
|
| 46 |
A |
539 |
516 |
1.31 |
|
|
|
| 47 |
A |
444 |
425 |
1.09 |
|
|
|
| 48 |
A |
440 |
421 |
2.06 |
|
|
|
| 49 |
A |
395 |
378 |
0.09 |
|
|
|
| 50 |
A |
335 |
321 |
11.90 |
|
|
|
| 51 |
A |
321 |
307 |
1.33 |
|
|
|
| 52 |
A |
252 |
241 |
24.79 |
|
|
|
| 53 |
A |
220 |
211 |
10.83 |
|
|
|
| 54 |
A |
154 |
147 |
0.32 |
|
|
|
Unscaled Zero Point Vibrational Energy (zpe) 41250.3 cm
-1
Scaled (by 0.9577) Zero Point Vibrational Energy (zpe) 39505.4 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Charges, Dipole, Quadrupole and Polarizability
Charges from optimized geometry at B1B95/6-31G(2df,p)
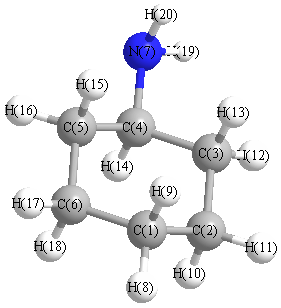
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.211 |
|
|
|
| 2 |
C |
-0.225 |
|
|
|
| 3 |
C |
-0.202 |
|
|
|
| 4 |
C |
0.033 |
|
|
|
| 5 |
C |
-0.224 |
|
|
|
| 6 |
C |
-0.222 |
|
|
|
| 7 |
N |
-0.556 |
|
|
|
| 8 |
H |
0.106 |
|
|
|
| 9 |
H |
0.106 |
|
|
|
| 10 |
H |
0.108 |
|
|
|
| 11 |
H |
0.107 |
|
|
|
| 12 |
H |
0.119 |
|
|
|
| 13 |
H |
0.097 |
|
|
|
| 14 |
H |
0.092 |
|
|
|
| 15 |
H |
0.097 |
|
|
|
| 16 |
H |
0.097 |
|
|
|
| 17 |
H |
0.106 |
|
|
|
| 18 |
H |
0.108 |
|
|
|
| 19 |
H |
0.233 |
|
|
|
| 20 |
H |
0.230 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.503 |
0.990 |
0.481 |
1.211 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-46.918 |
-3.605 |
-1.610 |
| y |
-3.605 |
-45.324 |
-0.630 |
| z |
-1.610 |
-0.630 |
-43.616 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
-2.448 |
-3.605 |
-1.610 |
| y |
-3.605 |
-0.057 |
-0.630 |
| z |
-1.610 |
-0.630 |
2.505 |
|
| Polar |
|---|
| 3z2-r2 | 5.010 |
|---|
| x2-y2 | -1.593 |
|---|
| xy | -3.605 |
|---|
| xz | -1.610 |
|---|
| yz | -0.630 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
11.414 |
-0.183 |
-0.118 |
| y |
-0.183 |
10.376 |
-0.104 |
| z |
-0.118 |
-0.104 |
9.314 |
<r2> (average value of r
2) Å
2
| <r2> |
229.909 |
| (<r2>)1/2 |
15.163 |