Vibrational Frequencies calculated at PBEPBE/cc-pVQZ
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3458 |
3458 |
0.18 |
|
|
|
| 2 |
A |
3378 |
3378 |
3.19 |
|
|
|
| 3 |
A |
3003 |
3003 |
38.36 |
|
|
|
| 4 |
A |
2995 |
2995 |
60.09 |
|
|
|
| 5 |
A |
2990 |
2990 |
45.02 |
|
|
|
| 6 |
A |
2989 |
2989 |
48.73 |
|
|
|
| 7 |
A |
2976 |
2976 |
57.32 |
|
|
|
| 8 |
A |
2945 |
2945 |
58.08 |
|
|
|
| 9 |
A |
2940 |
2940 |
10.11 |
|
|
|
| 10 |
A |
2935 |
2935 |
28.55 |
|
|
|
| 11 |
A |
2931 |
2931 |
37.27 |
|
|
|
| 12 |
A |
2920 |
2920 |
15.13 |
|
|
|
| 13 |
A |
2911 |
2911 |
1.45 |
|
|
|
| 14 |
A |
1599 |
1599 |
26.45 |
|
|
|
| 15 |
A |
1449 |
1449 |
2.27 |
|
|
|
| 16 |
A |
1436 |
1436 |
12.93 |
|
|
|
| 17 |
A |
1431 |
1431 |
5.39 |
|
|
|
| 18 |
A |
1430 |
1430 |
1.55 |
|
|
|
| 19 |
A |
1423 |
1423 |
1.18 |
|
|
|
| 20 |
A |
1366 |
1366 |
4.16 |
|
|
|
| 21 |
A |
1329 |
1329 |
2.12 |
|
|
|
| 22 |
A |
1325 |
1325 |
2.76 |
|
|
|
| 23 |
A |
1324 |
1324 |
0.14 |
|
|
|
| 24 |
A |
1315 |
1315 |
1.38 |
|
|
|
| 25 |
A |
1313 |
1313 |
0.83 |
|
|
|
| 26 |
A |
1284 |
1284 |
0.39 |
|
|
|
| 27 |
A |
1258 |
1258 |
0.90 |
|
|
|
| 28 |
A |
1244 |
1244 |
2.26 |
|
|
|
| 29 |
A |
1237 |
1237 |
1.59 |
|
|
|
| 30 |
A |
1185 |
1185 |
3.22 |
|
|
|
| 31 |
A |
1157 |
1157 |
1.13 |
|
|
|
| 32 |
A |
1096 |
1096 |
8.24 |
|
|
|
| 33 |
A |
1081 |
1081 |
3.10 |
|
|
|
| 34 |
A |
1064 |
1064 |
1.33 |
|
|
|
| 35 |
A |
1038 |
1038 |
0.17 |
|
|
|
| 36 |
A |
1017 |
1017 |
5.09 |
|
|
|
| 37 |
A |
1012 |
1012 |
0.26 |
|
|
|
| 38 |
A |
958 |
958 |
2.05 |
|
|
|
| 39 |
A |
907 |
907 |
16.85 |
|
|
|
| 40 |
A |
877 |
877 |
29.95 |
|
|
|
| 41 |
A |
860 |
860 |
16.26 |
|
|
|
| 42 |
A |
833 |
833 |
0.81 |
|
|
|
| 43 |
A |
831 |
831 |
102.76 |
|
|
|
| 44 |
A |
766 |
766 |
2.33 |
|
|
|
| 45 |
A |
763 |
763 |
2.35 |
|
|
|
| 46 |
A |
534 |
534 |
1.33 |
|
|
|
| 47 |
A |
444 |
444 |
0.87 |
|
|
|
| 48 |
A |
437 |
437 |
2.03 |
|
|
|
| 49 |
A |
394 |
394 |
0.16 |
|
|
|
| 50 |
A |
331 |
331 |
10.30 |
|
|
|
| 51 |
A |
321 |
321 |
1.05 |
|
|
|
| 52 |
A |
239 |
239 |
15.12 |
|
|
|
| 53 |
A |
210 |
210 |
18.27 |
|
|
|
| 54 |
A |
155 |
155 |
0.30 |
|
|
|
Unscaled Zero Point Vibrational Energy (zpe) 39820.7 cm
-1
Scaled (by 1) Zero Point Vibrational Energy (zpe) 39820.7 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Charges, Dipole, Quadrupole and Polarizability
Charges from optimized geometry at PBEPBE/cc-pVQZ
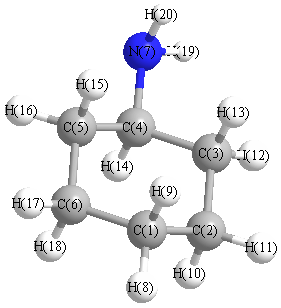
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.169 |
|
|
|
| 2 |
C |
-0.164 |
|
|
|
| 3 |
C |
-0.152 |
|
|
|
| 4 |
C |
0.068 |
|
|
|
| 5 |
C |
-0.144 |
|
|
|
| 6 |
C |
-0.178 |
|
|
|
| 7 |
N |
-0.383 |
|
|
|
| 8 |
H |
0.104 |
|
|
|
| 9 |
H |
0.063 |
|
|
|
| 10 |
H |
0.062 |
|
|
|
| 11 |
H |
0.106 |
|
|
|
| 12 |
H |
0.108 |
|
|
|
| 13 |
H |
0.041 |
|
|
|
| 14 |
H |
0.089 |
|
|
|
| 15 |
H |
0.042 |
|
|
|
| 16 |
H |
0.082 |
|
|
|
| 17 |
H |
0.105 |
|
|
|
| 18 |
H |
0.061 |
|
|
|
| 19 |
H |
0.132 |
|
|
|
| 20 |
H |
0.129 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.602 |
0.974 |
0.423 |
1.221 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-48.605 |
-3.648 |
-1.494 |
| y |
-3.648 |
-46.911 |
-0.730 |
| z |
-1.494 |
-0.730 |
-45.044 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
-2.628 |
-3.648 |
-1.494 |
| y |
-3.648 |
-0.086 |
-0.730 |
| z |
-1.494 |
-0.730 |
2.714 |
|
| Polar |
|---|
| 3z2-r2 | 5.428 |
|---|
| x2-y2 | -1.694 |
|---|
| xy | -3.648 |
|---|
| xz | -1.494 |
|---|
| yz | -0.730 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
13.665 |
-0.090 |
-0.132 |
| y |
-0.090 |
12.498 |
-0.002 |
| z |
-0.132 |
-0.002 |
10.924 |
<r2> (average value of r
2) Å
2
| <r2> |
234.195 |
| (<r2>)1/2 |
15.303 |