Jump to
S1C2
Vibrational Frequencies calculated at B2PLYP=FULLultrafine/TZVP
Geometric Data calculated at B2PLYP=FULLultrafine/TZVP
Electronic energy levels
Charges, Dipole, Quadrupole and Polarizability
Jump to
S1C1
Energy calculated at B2PLYP=FULLultrafine/TZVP
| | hartrees |
|---|
| Energy at 0K | -194.240595 |
| Energy at 298.15K | |
| HF Energy | -194.003501 |
| Nuclear repulsion energy | 132.746270 |
The energy at 298.15K was derived from the energy at 0K
and an integrated heat capacity that used the calculated vibrational frequencies.
Vibrational Frequencies calculated at B2PLYP=FULLultrafine/TZVP
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3851 |
3851 |
28.27 |
110.87 |
0.27 |
0.43 |
| 2 |
A |
3150 |
3150 |
24.19 |
37.49 |
0.74 |
0.85 |
| 3 |
A |
3123 |
3123 |
51.14 |
55.32 |
0.55 |
0.71 |
| 4 |
A |
3096 |
3096 |
21.30 |
83.81 |
0.74 |
0.85 |
| 5 |
A |
3061 |
3061 |
53.42 |
156.13 |
0.06 |
0.11 |
| 6 |
A |
3057 |
3057 |
34.70 |
120.51 |
0.05 |
0.09 |
| 7 |
A |
3042 |
3042 |
28.80 |
92.84 |
0.69 |
0.82 |
| 8 |
A |
3007 |
3007 |
61.48 |
120.22 |
0.10 |
0.19 |
| 9 |
A |
1540 |
1540 |
3.29 |
5.24 |
0.67 |
0.80 |
| 10 |
A |
1526 |
1526 |
6.98 |
5.40 |
0.75 |
0.86 |
| 11 |
A |
1512 |
1512 |
8.18 |
11.82 |
0.75 |
0.86 |
| 12 |
A |
1493 |
1493 |
2.58 |
12.31 |
0.74 |
0.85 |
| 13 |
A |
1469 |
1469 |
4.14 |
2.44 |
0.20 |
0.33 |
| 14 |
A |
1432 |
1432 |
3.48 |
0.70 |
0.75 |
0.85 |
| 15 |
A |
1400 |
1400 |
1.60 |
1.29 |
0.74 |
0.85 |
| 16 |
A |
1338 |
1338 |
14.33 |
11.89 |
0.73 |
0.84 |
| 17 |
A |
1289 |
1289 |
0.19 |
6.78 |
0.75 |
0.86 |
| 18 |
A |
1256 |
1256 |
38.42 |
3.12 |
0.64 |
0.78 |
| 19 |
A |
1176 |
1176 |
5.53 |
0.94 |
0.13 |
0.23 |
| 20 |
A |
1126 |
1126 |
5.01 |
4.11 |
0.58 |
0.73 |
| 21 |
A |
1078 |
1078 |
45.77 |
3.46 |
0.63 |
0.77 |
| 22 |
A |
991 |
991 |
51.01 |
4.56 |
0.75 |
0.85 |
| 23 |
A |
939 |
939 |
2.78 |
0.44 |
0.75 |
0.85 |
| 24 |
A |
875 |
875 |
1.51 |
10.54 |
0.14 |
0.25 |
| 25 |
A |
782 |
782 |
1.27 |
0.46 |
0.67 |
0.80 |
| 26 |
A |
482 |
482 |
9.34 |
0.26 |
0.57 |
0.72 |
| 27 |
A |
330 |
330 |
5.82 |
0.36 |
0.33 |
0.49 |
| 28 |
A |
242 |
242 |
91.79 |
2.22 |
0.74 |
0.85 |
| 29 |
A |
227 |
227 |
26.52 |
0.48 |
0.72 |
0.84 |
| 30 |
A |
141 |
141 |
12.88 |
0.14 |
0.66 |
0.79 |
Unscaled Zero Point Vibrational Energy (zpe) 24014.9 cm
-1
Scaled (by 1) Zero Point Vibrational Energy (zpe) 24014.9 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Geometric Data calculated at B2PLYP=FULLultrafine/TZVP
Point Group is C1
Cartesians (Å)
| Atom |
x (Å) |
y (Å) |
z (Å) |
|---|
| C1 |
-1.536 |
-0.518 |
0.128 |
| C2 |
-0.635 |
0.640 |
-0.289 |
| C3 |
0.762 |
0.547 |
0.291 |
| O4 |
1.384 |
-0.636 |
-0.215 |
| H5 |
-2.533 |
-0.414 |
-0.303 |
| H6 |
-1.645 |
-0.554 |
1.214 |
| H7 |
-1.120 |
-1.469 |
-0.200 |
| H8 |
-0.554 |
0.681 |
-1.378 |
| H9 |
-1.070 |
1.589 |
0.034 |
| H10 |
0.704 |
0.512 |
1.386 |
| H11 |
1.341 |
1.434 |
0.011 |
| H12 |
2.255 |
-0.714 |
0.184 |
Atom - Atom Distances (Å)
| |
C1 |
C2 |
C3 |
O4 |
H5 |
H6 |
H7 |
H8 |
H9 |
H10 |
H11 |
H12 |
| C1 | | 1.5255 | 2.5389 | 2.9431 | 1.0901 | 1.0919 | 1.0883 | 2.1606 | 2.1600 | 2.7687 | 3.4787 | 3.7972 |
C2 | 1.5255 | | 1.5161 | 2.3895 | 2.1705 | 2.1689 | 2.1656 | 1.0920 | 1.0931 | 2.1487 | 2.1504 | 3.2267 | C3 | 2.5389 | 1.5161 | | 1.4292 | 3.4833 | 2.8031 | 2.8018 | 2.1301 | 2.1239 | 1.0966 | 1.0952 | 1.9575 | O4 | 2.9431 | 2.3895 | 1.4292 | | 3.9241 | 3.3500 | 2.6395 | 2.6158 | 3.3221 | 2.0842 | 2.0825 | 0.9614 | H5 | 1.0901 | 2.1705 | 3.4833 | 3.9241 | | 1.7628 | 1.7658 | 2.5035 | 2.5025 | 3.7666 | 4.3029 | 4.8219 | H6 | 1.0919 | 2.1689 | 2.8031 | 3.3500 | 1.7628 | | 1.7632 | 3.0706 | 2.5131 | 2.5855 | 3.7830 | 4.0368 | H7 | 1.0883 | 2.1656 | 2.8018 | 2.6395 | 1.7658 | 1.7632 | | 2.5158 | 3.0671 | 3.1255 | 3.8115 | 3.4801 | H8 | 2.1606 | 1.0920 | 2.1301 | 2.6158 | 2.5035 | 3.0706 | 2.5158 | | 1.7556 | 3.0414 | 2.4670 | 3.5041 | H9 | 2.1600 | 1.0931 | 2.1239 | 3.3221 | 2.5025 | 2.5131 | 3.0671 | 1.7556 | | 2.4775 | 2.4161 | 4.0482 | H10 | 2.7687 | 2.1487 | 1.0966 | 2.0842 | 3.7666 | 2.5855 | 3.1255 | 3.0414 | 2.4775 | | 1.7734 | 2.3141 | H11 | 3.4787 | 2.1504 | 1.0952 | 2.0825 | 4.3029 | 3.7830 | 3.8115 | 2.4670 | 2.4161 | 1.7734 | | 2.3414 | H12 | 3.7972 | 3.2267 | 1.9575 | 0.9614 | 4.8219 | 4.0368 | 3.4801 | 3.5041 | 4.0482 | 2.3141 | 2.3414 | |
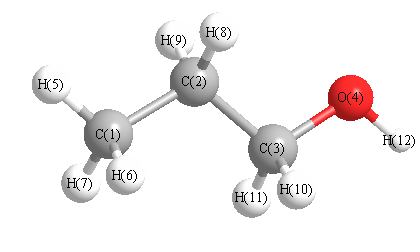 More geometry information
More geometry information
Calculated Bond Angles
| atom1 |
atom2 |
atom3 |
angle |
|
atom1 |
atom2 |
atom3 |
angle |
| C1 |
C2 |
C3 |
113.176 |
|
C1 |
C2 |
H8 |
110.165 |
| C1 |
C2 |
H9 |
110.055 |
|
C2 |
C1 |
H5 |
111.069 |
| C2 |
C1 |
H6 |
110.832 |
|
C2 |
C1 |
H7 |
110.787 |
| C2 |
C3 |
O4 |
108.407 |
|
C2 |
C3 |
H10 |
109.603 |
| C2 |
C3 |
H11 |
109.821 |
|
C3 |
C2 |
H8 |
108.417 |
| C3 |
C2 |
H9 |
107.879 |
|
C3 |
O4 |
H12 |
108.345 |
| O4 |
C3 |
H10 |
110.518 |
|
O4 |
C3 |
H11 |
110.470 |
| H5 |
C1 |
H6 |
107.776 |
|
H5 |
C1 |
H7 |
108.303 |
| H6 |
C1 |
H7 |
107.950 |
|
H8 |
C2 |
H9 |
106.927 |
| H10 |
C3 |
H11 |
108.018 |
|
Electronic energy levels
Charges, Dipole, Quadrupole and Polarizability
Charges from optimized geometry at B2PLYP=FULLultrafine/TZVP
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.337 |
|
|
|
| 2 |
C |
-0.144 |
|
|
|
| 3 |
C |
-0.096 |
|
|
|
| 4 |
O |
-0.390 |
|
|
|
| 5 |
H |
0.112 |
|
|
|
| 6 |
H |
0.089 |
|
|
|
| 7 |
H |
0.128 |
|
|
|
| 8 |
H |
0.102 |
|
|
|
| 9 |
H |
0.093 |
|
|
|
| 10 |
H |
0.086 |
|
|
|
| 11 |
H |
0.096 |
|
|
|
| 12 |
H |
0.261 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.721 |
1.036 |
1.046 |
1.639 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-23.215 |
-0.084 |
2.099 |
| y |
-0.084 |
-27.722 |
-0.598 |
| z |
2.099 |
-0.598 |
-27.286 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
4.288 |
-0.084 |
2.099 |
| y |
-0.084 |
-2.471 |
-0.598 |
| z |
2.099 |
-0.598 |
-1.817 |
|
| Polar |
|---|
| 3z2-r2 | -3.634 |
|---|
| x2-y2 | 4.506 |
|---|
| xy | -0.084 |
|---|
| xz | 2.099 |
|---|
| yz | -0.598 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
6.651 |
0.047 |
0.094 |
| y |
0.047 |
6.032 |
0.016 |
| z |
0.094 |
0.016 |
5.705 |
<r2> (average value of r
2) Å
2
| <r2> |
95.381 |
| (<r2>)1/2 |
9.766 |
