Jump to
S1C2
Vibrational Frequencies calculated at B2PLYP=FULLultrafine/6-311G*
Geometric Data calculated at B2PLYP=FULLultrafine/6-311G*
Electronic energy levels
Charges, Dipole, Quadrupole and Polarizability
Jump to
S1C1
Energy calculated at B2PLYP=FULLultrafine/6-311G*
| | hartrees |
|---|
| Energy at 0K | -194.192954 |
| Energy at 298.15K | |
| HF Energy | -193.969793 |
| Nuclear repulsion energy | 132.856410 |
The energy at 298.15K was derived from the energy at 0K
and an integrated heat capacity that used the calculated vibrational frequencies.
Vibrational Frequencies calculated at B2PLYP=FULLultrafine/6-311G*
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3833 |
3833 |
12.66 |
111.97 |
0.32 |
0.48 |
| 2 |
A |
3155 |
3155 |
28.89 |
40.89 |
0.73 |
0.85 |
| 3 |
A |
3123 |
3123 |
60.48 |
58.95 |
0.59 |
0.74 |
| 4 |
A |
3097 |
3097 |
25.54 |
91.06 |
0.73 |
0.85 |
| 5 |
A |
3061 |
3061 |
37.92 |
176.56 |
0.04 |
0.07 |
| 6 |
A |
3058 |
3058 |
52.54 |
64.23 |
0.15 |
0.26 |
| 7 |
A |
3034 |
3034 |
49.58 |
95.78 |
0.75 |
0.86 |
| 8 |
A |
3001 |
3001 |
71.42 |
108.18 |
0.13 |
0.23 |
| 9 |
A |
1557 |
1557 |
2.41 |
5.25 |
0.70 |
0.83 |
| 10 |
A |
1545 |
1545 |
8.74 |
6.55 |
0.75 |
0.86 |
| 11 |
A |
1530 |
1530 |
9.10 |
15.28 |
0.75 |
0.86 |
| 12 |
A |
1510 |
1510 |
3.15 |
13.39 |
0.73 |
0.84 |
| 13 |
A |
1484 |
1484 |
2.86 |
2.55 |
0.19 |
0.32 |
| 14 |
A |
1441 |
1441 |
5.19 |
1.31 |
0.67 |
0.80 |
| 15 |
A |
1405 |
1405 |
0.81 |
1.25 |
0.72 |
0.84 |
| 16 |
A |
1352 |
1352 |
23.74 |
12.55 |
0.74 |
0.85 |
| 17 |
A |
1293 |
1293 |
0.43 |
7.16 |
0.75 |
0.86 |
| 18 |
A |
1272 |
1272 |
47.01 |
4.38 |
0.59 |
0.75 |
| 19 |
A |
1181 |
1181 |
6.52 |
0.79 |
0.23 |
0.37 |
| 20 |
A |
1134 |
1134 |
5.88 |
4.10 |
0.62 |
0.76 |
| 21 |
A |
1088 |
1088 |
48.04 |
2.83 |
0.66 |
0.79 |
| 22 |
A |
1001 |
1001 |
43.67 |
4.43 |
0.75 |
0.86 |
| 23 |
A |
942 |
942 |
2.71 |
0.36 |
0.74 |
0.85 |
| 24 |
A |
881 |
881 |
2.10 |
9.56 |
0.16 |
0.28 |
| 25 |
A |
785 |
785 |
0.87 |
0.54 |
0.61 |
0.76 |
| 26 |
A |
486 |
486 |
9.60 |
0.24 |
0.45 |
0.62 |
| 27 |
A |
334 |
334 |
13.08 |
0.46 |
0.51 |
0.68 |
| 28 |
A |
270 |
270 |
125.49 |
3.93 |
0.73 |
0.85 |
| 29 |
A |
232 |
232 |
4.00 |
0.08 |
0.60 |
0.75 |
| 30 |
A |
149 |
149 |
7.85 |
0.14 |
0.74 |
0.85 |
Unscaled Zero Point Vibrational Energy (zpe) 24116.6 cm
-1
Scaled (by 1) Zero Point Vibrational Energy (zpe) 24116.6 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Geometric Data calculated at B2PLYP=FULLultrafine/6-311G*
Point Group is C1
Cartesians (Å)
| Atom |
x (Å) |
y (Å) |
z (Å) |
|---|
| C1 |
-1.531 |
-0.517 |
0.128 |
| C2 |
-0.632 |
0.646 |
-0.287 |
| C3 |
0.768 |
0.548 |
0.291 |
| O4 |
1.370 |
-0.640 |
-0.216 |
| H5 |
-2.522 |
-0.431 |
-0.322 |
| H6 |
-1.661 |
-0.545 |
1.214 |
| H7 |
-1.097 |
-1.467 |
-0.182 |
| H8 |
-0.549 |
0.684 |
-1.377 |
| H9 |
-1.070 |
1.598 |
0.031 |
| H10 |
0.717 |
0.521 |
1.388 |
| H11 |
1.353 |
1.431 |
0.004 |
| H12 |
2.241 |
-0.733 |
0.178 |
Atom - Atom Distances (Å)
| |
C1 |
C2 |
C3 |
O4 |
H5 |
H6 |
H7 |
H8 |
H9 |
H10 |
H11 |
H12 |
| C1 | | 1.5272 | 2.5391 | 2.9241 | 1.0918 | 1.0935 | 1.0894 | 2.1618 | 2.1664 | 2.7784 | 3.4828 | 3.7784 |
C2 | 1.5272 | | 1.5177 | 2.3800 | 2.1755 | 2.1744 | 2.1652 | 1.0939 | 1.0949 | 2.1546 | 2.1545 | 3.2200 | C3 | 2.5391 | 1.5177 | | 1.4243 | 3.4870 | 2.8187 | 2.7854 | 2.1291 | 2.1323 | 1.0992 | 1.0979 | 1.9548 | O4 | 2.9241 | 2.3800 | 1.4243 | | 3.8993 | 3.3521 | 2.6018 | 2.6038 | 3.3192 | 2.0849 | 2.0827 | 0.9603 | H5 | 1.0918 | 2.1755 | 3.4870 | 3.8993 | | 1.7644 | 1.7679 | 2.5004 | 2.5195 | 3.7846 | 4.3117 | 4.7986 | H6 | 1.0935 | 2.1744 | 2.8187 | 3.3521 | 1.7644 | | 1.7647 | 3.0756 | 2.5177 | 2.6115 | 3.8017 | 4.0406 | H7 | 1.0894 | 2.1652 | 2.7854 | 2.6018 | 1.7679 | 1.7647 | | 2.5206 | 3.0716 | 3.1153 | 3.7992 | 3.4359 | H8 | 2.1618 | 1.0939 | 2.1291 | 2.6038 | 2.5004 | 3.0756 | 2.5206 | | 1.7576 | 3.0456 | 2.4658 | 3.4936 | H9 | 2.1664 | 1.0949 | 2.1323 | 3.3192 | 2.5195 | 2.5177 | 3.0716 | 1.7576 | | 2.4886 | 2.4283 | 4.0510 | H10 | 2.7784 | 2.1546 | 1.0992 | 2.0849 | 3.7846 | 2.6115 | 3.1153 | 3.0456 | 2.4886 | | 1.7753 | 2.3147 | H11 | 3.4828 | 2.1545 | 1.0979 | 2.0827 | 4.3117 | 3.8017 | 3.7992 | 2.4658 | 2.4283 | 1.7753 | | 2.3460 | H12 | 3.7784 | 3.2200 | 1.9548 | 0.9603 | 4.7986 | 4.0406 | 3.4359 | 3.4936 | 4.0510 | 2.3147 | 2.3460 | |
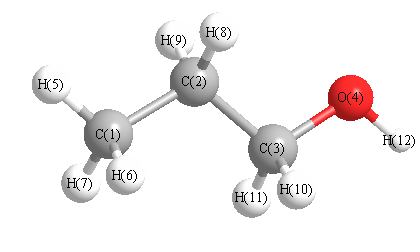 More geometry information
More geometry information
Calculated Bond Angles
| atom1 |
atom2 |
atom3 |
angle |
|
atom1 |
atom2 |
atom3 |
angle |
| C1 |
C2 |
C3 |
112.997 |
|
C1 |
C2 |
H8 |
110.026 |
| C1 |
C2 |
H9 |
110.332 |
|
C2 |
C1 |
H5 |
111.241 |
| C2 |
C1 |
H6 |
111.048 |
|
C2 |
C1 |
H7 |
110.567 |
| C2 |
C3 |
O4 |
107.947 |
|
C2 |
C3 |
H10 |
109.798 |
| C2 |
C3 |
H11 |
109.871 |
|
C3 |
C2 |
H8 |
108.124 |
| C3 |
C2 |
H9 |
108.309 |
|
C3 |
O4 |
H12 |
108.554 |
| O4 |
C3 |
H10 |
110.759 |
|
O4 |
C3 |
H11 |
110.667 |
| H5 |
C1 |
H6 |
107.681 |
|
H5 |
C1 |
H7 |
108.289 |
| H6 |
C1 |
H7 |
107.882 |
|
H8 |
C2 |
H9 |
106.833 |
| H10 |
C3 |
H11 |
107.802 |
|
Electronic energy levels
Charges, Dipole, Quadrupole and Polarizability
Charges from optimized geometry at B2PLYP=FULLultrafine/6-311G*
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.626 |
|
|
|
| 2 |
C |
-0.427 |
|
|
|
| 3 |
C |
-0.172 |
|
|
|
| 4 |
O |
-0.580 |
|
|
|
| 5 |
H |
0.208 |
|
|
|
| 6 |
H |
0.198 |
|
|
|
| 7 |
H |
0.230 |
|
|
|
| 8 |
H |
0.213 |
|
|
|
| 9 |
H |
0.199 |
|
|
|
| 10 |
H |
0.178 |
|
|
|
| 11 |
H |
0.188 |
|
|
|
| 12 |
H |
0.390 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.895 |
0.945 |
1.046 |
1.670 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-22.873 |
-0.298 |
2.026 |
| y |
-0.298 |
-27.542 |
-0.583 |
| z |
2.026 |
-0.583 |
-27.163 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
4.479 |
-0.298 |
2.026 |
| y |
-0.298 |
-2.524 |
-0.583 |
| z |
2.026 |
-0.583 |
-1.955 |
|
| Polar |
|---|
| 3z2-r2 | -3.910 |
|---|
| x2-y2 | 4.669 |
|---|
| xy | -0.298 |
|---|
| xz | 2.026 |
|---|
| yz | -0.583 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
6.286 |
0.037 |
0.140 |
| y |
0.037 |
5.620 |
0.008 |
| z |
0.140 |
0.008 |
5.317 |
<r2> (average value of r
2) Å
2
| <r2> |
94.930 |
| (<r2>)1/2 |
9.743 |
