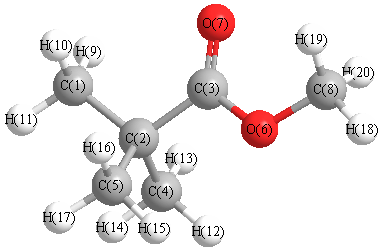
Vibrational Frequencies calculated at B3LYP/aug-cc-pVTZ
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3148 |
3046 |
13.77 |
|
|
|
| 2 |
A |
3117 |
3016 |
19.35 |
|
|
|
| 3 |
A |
3113 |
3011 |
35.73 |
|
|
|
| 4 |
A |
3112 |
3011 |
23.37 |
|
|
|
| 5 |
A |
3109 |
3008 |
0.00 |
|
|
|
| 6 |
A |
3102 |
3001 |
34.26 |
|
|
|
| 7 |
A |
3087 |
2986 |
24.17 |
|
|
|
| 8 |
A |
3085 |
2985 |
10.76 |
|
|
|
| 9 |
A |
3049 |
2950 |
33.83 |
|
|
|
| 10 |
A |
3039 |
2940 |
26.54 |
|
|
|
| 11 |
A |
3029 |
2931 |
12.06 |
|
|
|
| 12 |
A |
3027 |
2929 |
30.88 |
|
|
|
| 13 |
A |
1775 |
1718 |
238.37 |
|
|
|
| 14 |
A |
1523 |
1473 |
25.82 |
|
|
|
| 15 |
A |
1505 |
1456 |
10.84 |
|
|
|
| 16 |
A |
1501 |
1452 |
7.23 |
|
|
|
| 17 |
A |
1498 |
1450 |
7.09 |
|
|
|
| 18 |
A |
1492 |
1443 |
0.06 |
|
|
|
| 19 |
A |
1488 |
1440 |
0.10 |
|
|
|
| 20 |
A |
1484 |
1436 |
8.59 |
|
|
|
| 21 |
A |
1480 |
1431 |
0.13 |
|
|
|
| 22 |
A |
1470 |
1422 |
7.45 |
|
|
|
| 23 |
A |
1434 |
1387 |
8.80 |
|
|
|
| 24 |
A |
1405 |
1359 |
4.91 |
|
|
|
| 25 |
A |
1400 |
1355 |
5.53 |
|
|
|
| 26 |
A |
1299 |
1257 |
56.02 |
|
|
|
| 27 |
A |
1249 |
1208 |
2.93 |
|
|
|
| 28 |
A |
1227 |
1188 |
2.43 |
|
|
|
| 29 |
A |
1210 |
1171 |
81.65 |
|
|
|
| 30 |
A |
1176 |
1137 |
280.52 |
|
|
|
| 31 |
A |
1173 |
1135 |
1.13 |
|
|
|
| 32 |
A |
1058 |
1023 |
16.47 |
|
|
|
| 33 |
A |
1052 |
1018 |
0.08 |
|
|
|
| 34 |
A |
999 |
966 |
8.00 |
|
|
|
| 35 |
A |
973 |
942 |
0.00 |
|
|
|
| 36 |
A |
947 |
916 |
1.93 |
|
|
|
| 37 |
A |
940 |
909 |
1.13 |
|
|
|
| 38 |
A |
867 |
839 |
13.02 |
|
|
|
| 39 |
A |
793 |
768 |
2.00 |
|
|
|
| 40 |
A |
780 |
755 |
4.73 |
|
|
|
| 41 |
A |
586 |
567 |
2.01 |
|
|
|
| 42 |
A |
494 |
478 |
2.52 |
|
|
|
| 43 |
A |
381 |
368 |
0.96 |
|
|
|
| 44 |
A |
363 |
351 |
9.71 |
|
|
|
| 45 |
A |
341 |
330 |
0.22 |
|
|
|
| 46 |
A |
319 |
309 |
6.69 |
|
|
|
| 47 |
A |
288 |
279 |
2.47 |
|
|
|
| 48 |
A |
253 |
244 |
1.34 |
|
|
|
| 49 |
A |
250 |
242 |
0.06 |
|
|
|
| 50 |
A |
202 |
196 |
0.34 |
|
|
|
| 51 |
A |
199 |
193 |
2.28 |
|
|
|
| 52 |
A |
133 |
129 |
0.78 |
|
|
|
| 53 |
A |
118 |
114 |
1.45 |
|
|
|
| 54 |
A |
29 |
28 |
0.93 |
|
|
|
Unscaled Zero Point Vibrational Energy (zpe) 38084.8 cm
-1
Scaled (by 0.9675) Zero Point Vibrational Energy (zpe) 36847.0 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Charges, Dipole, Quadrupole and Polarizability
Charges from optimized geometry at B3LYP/aug-cc-pVTZ
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-1.089 |
|
|
|
| 2 |
C |
2.163 |
|
|
|
| 3 |
C |
0.631 |
|
|
|
| 4 |
C |
-1.077 |
|
|
|
| 5 |
C |
-1.076 |
|
|
|
| 6 |
O |
-0.349 |
|
|
|
| 7 |
O |
-0.931 |
|
|
|
| 8 |
C |
-0.624 |
|
|
|
| 9 |
H |
0.155 |
|
|
|
| 10 |
H |
0.155 |
|
|
|
| 11 |
H |
0.163 |
|
|
|
| 12 |
H |
0.187 |
|
|
|
| 13 |
H |
0.186 |
|
|
|
| 14 |
H |
0.151 |
|
|
|
| 15 |
H |
0.186 |
|
|
|
| 16 |
H |
0.186 |
|
|
|
| 17 |
H |
0.151 |
|
|
|
| 18 |
H |
0.245 |
|
|
|
| 19 |
H |
0.294 |
|
|
|
| 20 |
H |
0.294 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
-0.067 |
-1.863 |
0.001 |
1.864 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-44.645 |
1.925 |
0.000 |
| y |
1.925 |
-56.001 |
0.000 |
| z |
0.000 |
0.000 |
-49.660 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
8.186 |
1.925 |
0.000 |
| y |
1.925 |
-8.848 |
0.000 |
| z |
0.000 |
0.000 |
0.662 |
|
| Polar |
|---|
| 3z2-r2 | 1.325 |
|---|
| x2-y2 | 11.356 |
|---|
| xy | 1.925 |
|---|
| xz | 0.000 |
|---|
| yz | 0.000 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
13.742 |
0.152 |
0.000 |
| y |
0.152 |
12.088 |
-0.000 |
| z |
0.000 |
-0.000 |
10.966 |
<r2> (average value of r
2) Å
2
| <r2> |
298.549 |
| (<r2>)1/2 |
17.279 |