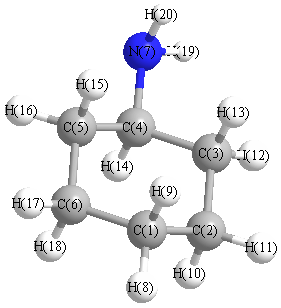
Vibrational Frequencies calculated at B3LYP/cc-pVQZ
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3549 |
3438 |
0.15 |
|
|
|
| 2 |
A |
3472 |
3364 |
1.43 |
|
|
|
| 3 |
A |
3064 |
2969 |
42.11 |
|
|
|
| 4 |
A |
3052 |
2957 |
77.77 |
|
|
|
| 5 |
A |
3048 |
2953 |
51.61 |
|
|
|
| 6 |
A |
3047 |
2952 |
54.70 |
|
|
|
| 7 |
A |
3036 |
2941 |
63.23 |
|
|
|
| 8 |
A |
3008 |
2914 |
68.62 |
|
|
|
| 9 |
A |
3002 |
2909 |
8.17 |
|
|
|
| 10 |
A |
2999 |
2905 |
40.08 |
|
|
|
| 11 |
A |
2996 |
2902 |
10.05 |
|
|
|
| 12 |
A |
2985 |
2892 |
12.94 |
|
|
|
| 13 |
A |
2980 |
2887 |
3.25 |
|
|
|
| 14 |
A |
1654 |
1602 |
28.54 |
|
|
|
| 15 |
A |
1509 |
1462 |
1.90 |
|
|
|
| 16 |
A |
1496 |
1449 |
10.97 |
|
|
|
| 17 |
A |
1491 |
1445 |
4.58 |
|
|
|
| 18 |
A |
1490 |
1443 |
1.20 |
|
|
|
| 19 |
A |
1483 |
1437 |
0.77 |
|
|
|
| 20 |
A |
1420 |
1376 |
4.52 |
|
|
|
| 21 |
A |
1382 |
1339 |
1.85 |
|
|
|
| 22 |
A |
1378 |
1335 |
3.36 |
|
|
|
| 23 |
A |
1377 |
1334 |
0.02 |
|
|
|
| 24 |
A |
1368 |
1325 |
0.89 |
|
|
|
| 25 |
A |
1367 |
1324 |
0.87 |
|
|
|
| 26 |
A |
1340 |
1298 |
0.40 |
|
|
|
| 27 |
A |
1303 |
1262 |
1.07 |
|
|
|
| 28 |
A |
1288 |
1248 |
2.34 |
|
|
|
| 29 |
A |
1281 |
1241 |
1.10 |
|
|
|
| 30 |
A |
1227 |
1189 |
3.01 |
|
|
|
| 31 |
A |
1198 |
1160 |
1.21 |
|
|
|
| 32 |
A |
1123 |
1088 |
10.76 |
|
|
|
| 33 |
A |
1110 |
1075 |
4.38 |
|
|
|
| 34 |
A |
1091 |
1057 |
0.02 |
|
|
|
| 35 |
A |
1074 |
1041 |
0.53 |
|
|
|
| 36 |
A |
1043 |
1011 |
6.93 |
|
|
|
| 37 |
A |
1035 |
1002 |
0.83 |
|
|
|
| 38 |
A |
984 |
954 |
1.89 |
|
|
|
| 39 |
A |
931 |
902 |
5.87 |
|
|
|
| 40 |
A |
905 |
877 |
39.31 |
|
|
|
| 41 |
A |
884 |
856 |
5.26 |
|
|
|
| 42 |
A |
851 |
824 |
47.42 |
|
|
|
| 43 |
A |
844 |
818 |
74.28 |
|
|
|
| 44 |
A |
792 |
767 |
0.42 |
|
|
|
| 45 |
A |
781 |
756 |
5.50 |
|
|
|
| 46 |
A |
553 |
535 |
1.16 |
|
|
|
| 47 |
A |
460 |
446 |
0.91 |
|
|
|
| 48 |
A |
452 |
437 |
1.85 |
|
|
|
| 49 |
A |
408 |
395 |
0.20 |
|
|
|
| 50 |
A |
342 |
332 |
10.28 |
|
|
|
| 51 |
A |
331 |
320 |
0.85 |
|
|
|
| 52 |
A |
245 |
237 |
16.83 |
|
|
|
| 53 |
A |
217 |
211 |
18.33 |
|
|
|
| 54 |
A |
159 |
154 |
0.28 |
|
|
|
Unscaled Zero Point Vibrational Energy (zpe) 40950.8 cm
-1
Scaled (by 0.9688) Zero Point Vibrational Energy (zpe) 39673.2 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Charges, Dipole, Quadrupole and Polarizability
Charges from optimized geometry at B3LYP/cc-pVQZ
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.152 |
|
|
|
| 2 |
C |
-0.143 |
|
|
|
| 3 |
C |
-0.150 |
|
|
|
| 4 |
C |
0.082 |
|
|
|
| 5 |
C |
-0.139 |
|
|
|
| 6 |
C |
-0.157 |
|
|
|
| 7 |
N |
-0.398 |
|
|
|
| 8 |
H |
0.092 |
|
|
|
| 9 |
H |
0.058 |
|
|
|
| 10 |
H |
0.058 |
|
|
|
| 11 |
H |
0.093 |
|
|
|
| 12 |
H |
0.103 |
|
|
|
| 13 |
H |
0.042 |
|
|
|
| 14 |
H |
0.092 |
|
|
|
| 15 |
H |
0.043 |
|
|
|
| 16 |
H |
0.075 |
|
|
|
| 17 |
H |
0.092 |
|
|
|
| 18 |
H |
0.057 |
|
|
|
| 19 |
H |
0.130 |
|
|
|
| 20 |
H |
0.125 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.596 |
0.995 |
0.413 |
1.231 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-48.592 |
-3.685 |
-1.453 |
| y |
-3.685 |
-46.938 |
-0.729 |
| z |
-1.453 |
-0.729 |
-44.894 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
-2.676 |
-3.685 |
-1.453 |
| y |
-3.685 |
-0.195 |
-0.729 |
| z |
-1.453 |
-0.729 |
2.871 |
|
| Polar |
|---|
| 3z2-r2 | 5.741 |
|---|
| x2-y2 | -1.654 |
|---|
| xy | -3.685 |
|---|
| xz | -1.453 |
|---|
| yz | -0.729 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
13.039 |
-0.087 |
-0.115 |
| y |
-0.087 |
11.959 |
-0.012 |
| z |
-0.115 |
-0.012 |
10.489 |
<r2> (average value of r
2) Å
2
| <r2> |
233.516 |
| (<r2>)1/2 |
15.281 |