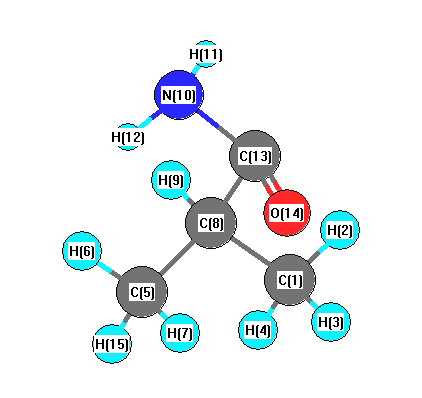
Vibrational Frequencies calculated at B3LYP/TZVP
| Mode Number |
Symmetry |
Frequency
(cm-1) |
Scaled Frequency
(cm-1) |
IR Intensities
(km mol-1) |
Raman Act
(Å4/u) |
Dep P |
Dep U |
|---|
| 1 |
A |
3722 |
3593 |
35.22 |
|
|
|
| 2 |
A |
3585 |
3461 |
34.34 |
|
|
|
| 3 |
A |
3117 |
3009 |
31.73 |
|
|
|
| 4 |
A |
3113 |
3005 |
2.65 |
|
|
|
| 5 |
A |
3097 |
2990 |
42.06 |
|
|
|
| 6 |
A |
3095 |
2988 |
17.51 |
|
|
|
| 7 |
A |
3037 |
2932 |
15.35 |
|
|
|
| 8 |
A |
3033 |
2929 |
33.36 |
|
|
|
| 9 |
A |
3018 |
2914 |
33.04 |
|
|
|
| 10 |
A |
1761 |
1701 |
296.03 |
|
|
|
| 11 |
A |
1622 |
1566 |
106.07 |
|
|
|
| 12 |
A |
1519 |
1466 |
16.01 |
|
|
|
| 13 |
A |
1506 |
1454 |
10.99 |
|
|
|
| 14 |
A |
1497 |
1445 |
2.39 |
|
|
|
| 15 |
A |
1491 |
1439 |
1.03 |
|
|
|
| 16 |
A |
1427 |
1377 |
32.08 |
|
|
|
| 17 |
A |
1403 |
1355 |
32.89 |
|
|
|
| 18 |
A |
1402 |
1354 |
4.57 |
|
|
|
| 19 |
A |
1338 |
1292 |
1.36 |
|
|
|
| 20 |
A |
1280 |
1236 |
119.77 |
|
|
|
| 21 |
A |
1199 |
1158 |
5.79 |
|
|
|
| 22 |
A |
1137 |
1098 |
6.32 |
|
|
|
| 23 |
A |
1120 |
1081 |
0.63 |
|
|
|
| 24 |
A |
1061 |
1024 |
7.77 |
|
|
|
| 25 |
A |
976 |
942 |
0.11 |
|
|
|
| 26 |
A |
942 |
909 |
1.52 |
|
|
|
| 27 |
A |
912 |
881 |
2.58 |
|
|
|
| 28 |
A |
773 |
746 |
4.44 |
|
|
|
| 29 |
A |
753 |
727 |
3.37 |
|
|
|
| 30 |
A |
620 |
598 |
5.33 |
|
|
|
| 31 |
A |
594 |
573 |
12.70 |
|
|
|
| 32 |
A |
479 |
463 |
2.74 |
|
|
|
| 33 |
A |
328 |
316 |
1.10 |
|
|
|
| 34 |
A |
290 |
280 |
5.40 |
|
|
|
| 35 |
A |
256 |
247 |
135.72 |
|
|
|
| 36 |
A |
243 |
235 |
4.45 |
|
|
|
| 37 |
A |
239 |
230 |
54.59 |
|
|
|
| 38 |
A |
215 |
208 |
7.20 |
|
|
|
| 39 |
A |
33 |
32 |
7.55 |
|
|
|
Unscaled Zero Point Vibrational Energy (zpe) 28615.4 cm
-1
Scaled (by 0.9654) Zero Point Vibrational Energy (zpe) 27625.3 cm
-1
See section
III.C.1 List or set vibrational scaling factors
to change the scale factors used here.
See section
III.C.2
Calculate a vibrational scaling factor for a given set of molecules
to determine the least squares best scaling factor.
Charges, Dipole, Quadrupole and Polarizability
Charges from optimized geometry at B3LYP/TZVP
Charges (e)
| Number |
Element |
Mulliken |
CHELPG |
AIM |
ESP |
| 1 |
C |
-0.367 |
|
|
|
| 2 |
H |
0.116 |
|
|
|
| 3 |
H |
0.144 |
|
|
|
| 4 |
H |
0.120 |
|
|
|
| 5 |
C |
-0.366 |
|
|
|
| 6 |
H |
0.116 |
|
|
|
| 7 |
H |
0.144 |
|
|
|
| 8 |
C |
-0.117 |
|
|
|
| 9 |
H |
0.106 |
|
|
|
| 10 |
N |
-0.387 |
|
|
|
| 11 |
H |
0.244 |
|
|
|
| 12 |
H |
0.239 |
|
|
|
| 13 |
C |
0.238 |
|
|
|
| 14 |
O |
-0.349 |
|
|
|
| 15 |
H |
0.120 |
|
|
|
Electric dipole moments
Electric dipole components in Debye
(What's a Debye? See section
VII.A.3)
| |
x |
y |
z |
Total |
| |
0.122 |
0.000 |
-3.732 |
3.734 |
| CHELPG |
|
|
|
|
| AIM |
|
|
|
|
| ESP |
|
|
|
|
Electric Quadrupole moment
Quadrupole components in D Å
| Primitive |
|---|
| | x | y | z |
|---|
| x |
-33.415 |
0.000 |
-4.902 |
| y |
0.000 |
-38.835 |
0.000 |
| z |
-4.902 |
0.000 |
-39.940 |
|
| Traceless |
|---|
| | x | y | z |
|---|
| x |
5.972 |
0.000 |
-4.902 |
| y |
0.000 |
-2.157 |
0.000 |
| z |
-4.902 |
0.000 |
-3.815 |
|
| Polar |
|---|
| 3z2-r2 | -7.630 |
|---|
| x2-y2 | 5.419 |
|---|
| xy | 0.000 |
|---|
| xz | -4.902 |
|---|
| yz | 0.000 |
|---|
|
Polarizabilities
Components of the polarizability tensor.
Units are
Å
3 (Angstrom cubed)
Change units.
| |
x |
y |
z |
| x |
9.604 |
0.000 |
-0.369 |
| y |
0.000 |
7.793 |
0.000 |
| z |
-0.369 |
0.000 |
8.690 |
<r2> (average value of r
2) Å
2
| <r2> |
173.823 |
| (<r2>)1/2 |
13.184 |