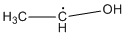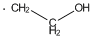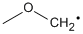Methods with standard basis sets
|
|
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
cc-pVQZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
aug-cc-pVQZ |
daug-cc-pVTZ |
| hartree fock |
HF |
103.8 a
341.9 b
116.4 c
0.0 d |
42.5 a
246.7 b
75.8 c
0.0 d |
42.5 a
246.7 b
75.8 c
0.0 d |
33.4 a
237.4 b
91.1 c
0.0 d |
26.2 a
233.4 b
65.4 c
0.0 d |
12.6 a
219.2 b
64.0 c
0.0 d |
6.2 a
210.4 b
59.7 c
0.0 d |
23.9 a
229.8 b
62.5 c
0.0 d |
8.1 a
212.8 b
60.0 c
0.0 d |
a
b
c |
|
NC
NC |
a
b
c |
a
b
c |
a |
a
b
c |
a
b
c |
a |
a
b
c |
| ROHF |
|
50.8 a
246.7 b
75.8 c
0.0 d |
50.8 a
246.7 b
75.8 c
0.0 d |
42.3 a
237.4 b
91.1 c
0.0 d |
36.5 a
233.4 b
65.4 c
0.0 d |
23.0 a
219.2 b
64.0 c
0.0 d |
30.5 a
225.3 b
61.4 c
0.0 d |
34.4 a
229.8 b
62.5 c
0.0 d |
18.7 a
212.8 b
60.0 c
0.0 d |
|
|
NC
NC |
a
b
c |
a
b
c |
a |
a
b
c |
a
b
c |
a |
|
| density functional |
LSDA |
a
b
c |
a
b
c |
a
b
c |
-20.4 a
196.9 b
33.6 c
0.0 d |
-28.2 a
188.9 b
13.9 c
0.0 d |
-38.8 a
177.9 b
14.0 c
0.0 d |
-43.2 a
167.7 b
15.4 c
0.0 d |
-29.5 a
186.3 b
16.6 c
0.0 d |
-44.8 a
170.0 b
14.1 c
0.0 d |
-43.2 a
171.7 b
7.3 c
0.0 d |
|
NC
NC |
-43.1 a
171.6 b
9.9 c
0.0 d |
-48.8 a
162.9 b
9.9 c
0.0 d |
|
-48.7 a
159.6 b
13.9 c
0.0 d |
-50.7 a
159.1 b
10.0 c
0.0 d |
|
|
| BLYP |
27.9 a
26.6 c
0.0 d |
a
b
c |
a
b
c |
-0.7 a
41.5 b
39.3 c
0.0 d |
a
b
c |
a
b
c |
-26.8 a
174.2 b
21.6 c
0.0 d |
-14.2 a
192.3 b
21.8 c
0.0 d |
-27.9 a
177.9 b
19.9 c
0.0 d |
a
b
c |
|
NC
NC |
a
b
c |
-31.6 a
171.6 b
16.5 c
0.0 d |
|
NC |
NC |
|
|
| B1B95 |
41.8 a
297.9 b
51.7 c
0.0 d |
a
b
c |
a
b
c |
0.2 a
213.3 b
53.5 c
0.0 d |
a
b
c |
a
b
c |
-28.4 a
181.5 b
27.9 c
0.0 d |
-12.9 a
200.5 b
29.7 c
0.0 d |
-28.4 a
184.2 b
27.2 c
0.0 d |
a
b
c |
|
NC
NC |
a
b
c |
a
b
c |
|
-32.8 a
174.8 b
27.9 c
0.0 d |
-34.7 a
174.1 b
21.4 c
0.0 d |
|
|
| B3LYP |
42.5 a
291.0 b
46.2 c
0.0 d |
a
b
c |
a
b
c |
1.5 a
209.7 b
48.1 c
0.0 d |
a
b
c |
a
b
c |
-27.0 a
177.7 b
24.9 c
0.0 d |
-12.8 a
196.2 b
26.1 c
0.0 d |
-27.5 a
180.8 b
23.8 c
0.0 d |
a
b
c |
|
NC
NC |
a
b
c |
-31.5 a
174.5 b
19.4 c
0.0 d |
-33.4 a
0.0 d |
-32.6 a
170.0 b
24.1 c
0.0 d |
-34.0 a
170.3 b
18.8 c
0.0 d |
-34.8 a
0.0 d |
|
| B3LYPultrafine |
|
NC |
|
|
a
b
c |
NC |
NC |
NC |
|
|
|
NC |
NC |
NC |
|
NC |
-34.0 a
2.0 b
18.8 c
0.0 d |
|
|
| B3PW91 |
44.0 a
294.1 b
53.7 c
0.0 d |
a
b
c |
a
b
c |
-1.4 a
208.1 b
52.1 c
0.0 d |
a
b
c |
a
b
c |
-30.5 a
175.8 b
26.7 c
0.0 d |
-15.1 a
194.4 b
28.7 c
0.0 d |
-30.6 a
178.3 b
26.2 c
0.0 d |
a
b
c |
|
NC
NC |
a
b
c |
-34.8 a
172.1 b
21.6 c
0.0 d |
|
NC |
NC |
|
|
| mPW1PW91 |
48.2 a
297.8 b
59.1 c
0.0 d |
a
b
c |
a
b
c |
0.2 a
209.9 b
55.2 c
0.0 d |
a
b
c |
a
b
c |
-29.8 a
177.0 b
28.4 c
0.0 d |
-13.6 a
196.2 b
30.6 c
0.0 d |
-29.4 a
179.7 b
28.1 c
0.0 d |
a
b
c |
|
NC
NC |
a
b
c |
-33.7 a
173.6 b
23.4 c
0.0 d |
|
-33.6 a
170.9 b
28.3 c
0.0 d |
-36.0 a
170.0 b
22.6 c
0.0 d |
|
|
| M06-2X |
42.7 a
290.8 b
47.6 c
0.0 d |
a
b
c |
a
b
c |
-1.4 a
206.8 b
49.4 c
0.0 d |
-16.1 a
193.1 b
21.1 c
0.0 d |
-27.7 a
181.1 b
20.3 c
0.0 d |
-34.0 a
171.0 b
18.4 c
0.0 d |
-18.5 a
189.3 b
20.0 c
0.0 d |
-33.8 a
173.1 b
17.4 c
0.0 d |
-34.0 a
175.0 b
0.0 d |
|
NC
NC |
-33.6 a
175.1 b
15.7 c
0.0 d |
-38.9 a
166.7 b
12.4 c
0.0 d |
|
-40.2 a
164.4 b
0.0 d |
-40.9 a
163.3 b
11.9 c
0.0 d |
|
|
| PBEPBE |
27.5 a
279.5 b
33.8 c
0.0 d |
a
b
c |
a
b
c |
-4.0 a
204.3 b
45.2 c
0.0 d |
a
b
c |
-25.5 a
183.6 b
25.1 c
0.0 d |
-31.0 a
173.0 b
25.0 c
0.0 d |
-16.7 a
191.9 b
25.9 c
0.0 d |
-31.6 a
176.3 b
23.7 c
0.0 d |
-30.1 a
177.6 b
18.7 c
0.0 d |
|
NC
NC |
-29.1 a
178.6 b
21.7 c
0.0 d |
-35.8 a
169.3 b
19.7 c
0.0 d |
|
-35.7 a
166.1 b
24.7 c
0.0 d |
-38.2 a
164.9 b
19.4 c
0.0 d |
|
|
| PBEPBEultrafine |
|
NC |
|
|
NC |
NC |
NC |
NC |
|
|
|
NC |
NC |
NC |
|
NC |
NC |
|
|
| PBE1PBE |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
|
NC |
NC |
NC |
|
NC |
NC |
|
|
| HSEh1PBE |
a
b
c |
a
b
c |
a
b
c |
0.7 a
210.2 b
55.9 c
0.0 d |
a
b
c |
a
b
c |
a |
a
b
c |
a
b
c |
a
b
c |
|
NC
NC |
a
b
c |
a
b
c |
|
a
b
c |
a
b
c |
|
|
| TPSSh |
NC |
NC |
NC |
NC |
a
b
c |
NC |
a
b
c |
NC |
NC |
a
b
c |
|
NC |
NC |
a
b
c |
NC |
NC |
NC |
NC |
|
| wB97X-D |
NC |
NC |
0.9 a
217.9 b
38.8 c
0.0 d |
NC |
-13.3 a
200.4 b
28.9 c
0.0 d |
NC |
-30.9 a
178.3 b
26.2 c
0.0 d |
NC |
-30.0 a
181.7 b
26.3 c
0.0 d |
NC |
|
-30.2 a
180.2 b
27.3 c
0.0 d |
-14.4 a
178.3 b
26.2 c
0.0 d |
-33.3 a
176.7 b
22.0 c
0.0 d |
NC |
NC |
-35.3 a
173.1 b
21.4 c
0.0 d |
NC |
|
| B97D3 |
|
b
c |
|
|
b
c |
|
172.2 b
23.2 c
0.0 d |
|
175.3 b
22.6 c
0.0 d |
|
-36.5 a
165.9 b
17.3 c
0.0 d |
172.9 b
23.8 c
0.0 d |
|
169.2 b
18.4 c
0.0 d |
|
|
165.1 b
18.1 c
0.0 d |
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
cc-pVQZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
aug-cc-pVQZ |
daug-cc-pVTZ |
|---|
| Moller Plesset perturbation |
MP2 |
79.9 a
316.9 b
87.0 c
0.0 d |
a
b
c |
a
b
c |
-13.0 a
189.3 b
42.7 c
0.0 d |
a
b
c |
a
b
c |
NC
NC |
a
b
c |
a
b
c |
a
b
c |
|
NC
NC |
a
b
c |
a
b
c |
a |
a
b
c |
a
c |
a |
|
| MP2=FULL |
80.1 a
317.1 b
87.3 c
0.0 d |
a
b
c |
a
b
c |
-13.1 a
189.3 b
42.6 c
0.0 d |
a
b
c |
a
b
c |
a
b
c |
a
b
c |
a
b
c |
a
b
c |
|
NC
NC |
a
b
c |
a
b
c |
a |
a
b
c |
a
b
c |
a |
|
| ROMP2 |
80.3 a
131.5 b
87.4 c
0.0 d |
a
b
c |
a
b
c |
-13.7 a
19.8 b
42.3 c
0.0 d |
a
c |
a
c |
a
c |
a
c |
a
c |
a
c |
|
NC |
a
c |
a
c |
|
a
c |
|
|
|
| MP3 |
|
|
|
|
a
b
c |
|
a |
|
|
|
|
NC |
NC |
NC |
|
|
|
|
|
| MP3=FULL |
|
NC |
NC |
NC |
a
b
c |
NC |
a
b
c |
NC |
NC |
NC |
|
NC |
NC |
NC |
|
NC |
|
|
|
| MP4 |
|
a
c |
|
|
a
c |
|
|
|
a
c |
|
|
NC |
NC |
a |
|
NC |
NC |
|
|
| MP4=FULL |
|
NC |
|
|
NC |
|
|
|
NC |
|
|
|
NC |
NC |
|
NC |
|
|
|
| B2PLYP |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
|
NC |
NC |
NC |
|
NC |
NC |
|
|
| B2PLYP=FULL |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
|
NC |
NC |
NC |
|
NC |
NC |
|
|
| Configuration interaction |
CID |
|
a
b
c |
a
b
c |
7.1 a
207.5 b
63.5 c
0.0 d |
a
b
c |
|
|
-6.1 a
200.8 b
37.2 c
0.0 d |
|
|
|
|
|
|
|
|
|
|
|
| CISD |
|
a
b
c |
a
b
c |
7.0 a
208.1 b
62.1 c
0.0 d |
a
b
c |
|
|
-5.7 a
201.9 b
36.9 c
0.0 d |
|
|
|
|
|
|
|
|
|
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
cc-pVQZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
aug-cc-pVQZ |
daug-cc-pVTZ |
|---|
| Quadratic configuration interaction |
QCISD |
|
a
b
c |
NC
NC
NC |
-1.5 a
198.1 b
51.0 c
0.0 d |
a
b
c |
-36.3 a
172.0 b
14.2 c
0.0 d |
a
b
c |
a
b
c |
-44.1 a
162.2 b
11.1 c
0.0 d |
a
c |
|
NC
NC |
a
b
c |
a
c |
|
a
c |
|
|
|
| QCISD(T) |
|
|
|
|
a
c |
|
|
NC |
|
|
|
NC |
-31.8 a
19.4 c
0.0 d |
a |
|
NC |
|
|
|
| QCISD(T)=FULL |
|
|
|
|
NC |
|
NC |
|
|
|
|
|
NC |
NC |
|
NC |
|
|
|
| Coupled Cluster |
CCD |
|
a
b
c |
a
b
c |
2.5 a
199.7 b
58.2 c
0.0 d |
a
b
c |
a
b
c |
a
b
c |
-11.3 a
192.8 b
33.4 c
0.0 d |
a
b
c |
a
c |
|
NC
NC |
a
b
c |
a
c |
|
a
c |
c |
|
|
| CCSD |
|
|
|
|
a
b
c |
|
|
|
|
NC |
|
NC |
a
b
c |
a
c |
|
NC |
|
|
|
| CCSD=FULL |
|
|
|
|
NC |
|
|
|
|
NC |
|
NC |
NC |
|
|
NC |
|
|
|
| CCSD(T) |
|
|
|
|
a
c |
NC |
|
|
|
|
|
NC |
a
c |
a |
|
a
c |
|
|
|
| CCSD(T)=FULL |
|
|
|
|
a
c |
|
|
|
|
|
|
NC |
a
c |
a |
|
a
c |
|
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
cc-pVQZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
aug-cc-pVQZ |
daug-cc-pVTZ |
|---|