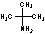
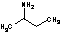
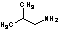
Methods with standard basis sets
|
|
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
cc-pV(T+d)Z |
daug-cc-pVTZ |
| hartree fock |
HF |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
23.3 a
0.0 b
10.0 c
16.7 d
16.4 e |
NC
NC
NC
NC |
31.6 a
0.0 b
10.1 c
16.9 d
17.3 e |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC |
30.7 a
0.0 b
9.7 c
16.3 d
16.4 e |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
0.0 b
9.1 c
15.9 d
15.6 e |
29.2 a
0.0 b
8.6 c
15.4 d
15.1 e |
| density functional |
LSDA |
NC |
NC
NC |
NC |
NC |
NC
NC |
NC
NC |
NC
NC |
NC
NC |
NC
NC |
NC
NC |
|
|
NC
NC |
NC
NC |
NC
NC |
NC |
NC |
|
| BLYP |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
17.9 a
0.0 b
9.8 c
16.9 d
15.4 e |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
|
NC |
NC
NC
NC
NC |
NC
NC |
NC
NC
NC |
NC |
e |
|
| B1B95 |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC |
|
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC |
NC |
0.0 b
9.8 c
17.1 d
20.0 e |
|
| B3LYP |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
30.0 a
0.0 b
10.6 c
17.7 d
18.7 e |
NC
NC
NC
NC |
NC
NC |
31.2 a
0.0 b
9.6 c
16.7 d
17.9 e |
NC
NC
NC
NC |
NC
NC
NC
NC |
33.4 a
0.0 b
8.9 c
15.6 d
17.7 e |
31.0 a
0.0 b
8.7 c
15.8 d
17.0 e |
0.0 b
8.9 c
16.1 d
17.0 e |
|
| B3LYPultrafine |
|
NC |
|
|
NC
NC
NC
NC |
NC |
NC |
NC |
|
|
|
NC |
NC |
NC |
NC |
NC
NC
NC
NC |
|
|
| B3PW91 |
9.6 a
0.0 b
11.8 c
16.4 d
11.7 e |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
32.7 a
0.0 b
11.0 c
17.9 d
19.1 e |
NC
NC
NC
NC |
|
NC |
NC
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC |
0.0 b
9.1 c
16.1 d
16.9 e |
|
| mPW1PW91 |
11.4 a
0.0 b
12.3 c
16.9 d
13.0 e |
NC
NC
NC
NC |
37.4 a
0.0 b
18.0 c
22.7 d
27.3 e |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
|
NC |
NC
NC
NC
NC |
33.0 a
0.0 b
9.8 c
16.8 d
18.4 e |
NC
NC
NC |
NC |
0.0 b
9.8 c
16.8 d
18.4 e |
|
| M06-2X |
NC |
NC |
44.2 a
0.0 b
21.5 c
25.4 d
37.0 e |
NC |
34.3 a
0.0 b
15.7 c
22.0 d
28.3 e |
NC |
NC |
NC |
NC |
NC |
|
NC |
NC |
NC |
NC |
NC |
|
|
| PBEPBE |
10.3 a
0.0 b
12.7 c
17.3 d
13.0 e |
NC
NC
NC
NC |
32.7 a
0.0 b
17.5 c
23.2 d
27.1 e |
30.9 a
0.0 b
12.1 c
19.0 d
20.6 e |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
|
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
35.8 a
0.0 b
9.4 c
16.2 d
19.3 e |
NC
NC |
0.0 b
9.9 c
17.0 d
19.0 e |
|
| PBEPBEultrafine |
|
NC |
|
|
NC
NC
NC
NC |
NC |
NC |
NC |
|
|
|
NC |
NC |
NC |
NC |
NC |
|
|
| PBE1PBE |
NC |
NC |
NC |
NC |
27.1 a
0.0 b
11.7 c
18.5 d
19.9 e |
NC |
NC |
NC |
NC |
NC |
|
NC |
NC |
NC |
NC |
NC |
|
|
| HSEh1PBE |
NC |
38.8 a
0.0 b
18.8 c
23.7 d
29.1 e |
NC |
NC |
27.0 a
0.0 b
11.7 c
18.6 d
20.1 e |
NC |
36.8 a
0.0 b
11.6 c
18.8 d
22.0 e |
NC |
NC |
NC |
|
NC |
NC |
34.9 a
0.0 b
10.7 c
17.7 d
20.3 e |
NC |
NC |
|
|
| TPSSh |
NC |
NC |
NC |
NC |
19.3 a
0.0 b
10.6 c
17.3 d
17.3 e |
NC |
28.1 a
0.0 b
10.2 c
17.1 d
18.8 e |
NC |
NC |
24.0 a
0.0 b
10.5 c
16.9 d
17.2 e |
|
NC |
NC |
26.5 a
0.0 b
9.4 c
16.3 d
17.4 e |
NC |
NC |
|
|
| wB97X-D |
NC |
NC |
42.2 a
0.0 b
19.4 c
23.0 d
29.3 e |
NC |
31.5 a
0.0 b
13.4 c
19.4 d
22.7 e |
NC |
41.1 a
0.0 b
13.6 c
19.9 d
24.4 e |
NC |
40.4 a
0.0 b
14.3 c
20.2 d
25.0 e |
NC |
|
41.0 a
0.0 b
13.1 c
19.3 d
23.7 e |
72.5 a
0.0 b
13.6 c
19.9 d
24.4 e |
39.0 a
0.0 b
12.6 c
18.8 d
23.1 e |
NC |
40.3 a
0.0 b
12.5 c
18.7 d
23.1 e |
|
|
| B97D3 |
|
0.0 b
19.8 c
24.8 d
30.1 e |
|
|
0.0 b
13.6 c
19.9 d
22.2 e |
|
0.0 b
13.2 c
19.9 d
24.0 e |
|
0.0 b
14.3 c
20.8 d
24.5 e |
|
38.0 a
0.0 b
12.6 c
18.8 d
23.3 e |
0.0 b
12.8 c
19.3 d
23.0 e |
|
0.0 b
12.5 c
19.0 d
22.6 e |
|
0.0 b
11.9 c
18.3 d
22.4 e |
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
cc-pV(T+d)Z |
daug-cc-pVTZ |
|---|
| Moller Plesset perturbation |
MP2 |
15.4 a
0.0 b
14.6 c
18.7 d
16.5 e |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
43.0 a
0.0 b
20.0 c
27.1 d
33.7 e |
NC
NC
NC
NC |
NC |
50.0 a
0.0 b
21.8 c
28.3 d
37.5 e |
NC
NC
NC
NC |
49.5 a
0.0 b
19.6 c
26.3 d
34.7 e |
|
53.6 a
0.0 b
20.1 c
26.9 d
36.5 e |
NC
NC
NC
NC |
NC
NC |
NC
NC |
NC |
NC |
|
| MP2=FULL |
NC
NC |
NC
NC |
NC
NC |
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
NC
NC
NC
NC |
53.7 a
0.0 b
21.1 c
27.5 d
36.0 e |
NC
NC |
|
NC |
NC
NC |
NC |
NC |
NC |
|
|
| MP3 |
|
|
|
|
NC
NC
NC
NC |
|
a |
|
|
|
|
NC |
NC |
NC |
|
|
|
|
| MP3=FULL |
|
NC |
NC |
NC |
39.8 a
0.0 b
17.4 c
24.3 d
29.2 e |
NC |
46.5 a
0.0 b
16.5 c
23.7 d
30.3 e |
NC |
NC |
NC |
|
NC |
NC |
|
|
|
|
|
| MP4 |
|
NC
NC |
|
|
NC
NC |
|
|
|
NC |
|
|
|
NC |
|
|
|
|
|
| MP4=FULL |
|
NC |
|
|
NC |
|
|
|
NC |
|
|
|
NC |
|
|
|
|
|
| B2PLYP |
NC |
NC |
NC |
NC |
29.3 a
0.0 b
13.9 c
20.9 d
23.2 e |
NC |
NC |
NC |
NC |
NC |
|
NC |
NC |
37.4 a
0.0 b
12.6 c
19.6 d
23.4 e |
NC |
NC |
|
|
| B2PLYP=FULL |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
|
NC |
NC |
NC |
NC |
NC |
|
|
| Configuration interaction |
CID |
|
NC
NC |
NC
NC |
NC
NC |
NC
NC
NC
NC |
|
|
NC
NC |
|
|
|
|
|
|
|
|
|
|
| CISD |
|
NC
NC |
NC
NC |
NC
NC |
NC
NC
NC |
|
|
NC
NC |
|
|
|
|
|
|
|
|
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
cc-pV(T+d)Z |
daug-cc-pVTZ |
|---|
| Quadratic configuration interaction |
QCISD |
NC |
NC
NC
NC
NC |
NC
NC |
NC
NC |
a
e |
NC |
NC |
NC
NC |
NC |
|
|
NC |
NC |
|
NC |
|
|
|
| QCISD(T) |
|
|
|
|
NC |
|
|
NC |
|
|
|
NC |
NC |
|
|
|
|
|
| QCISD(T)=FULL |
|
|
|
|
NC |
|
NC |
|
|
|
|
|
NC |
|
|
|
|
|
| Coupled Cluster |
CCD |
NC |
NC
NC |
NC
NC |
NC
NC |
NC
NC
NC
NC |
NC
NC |
NC
NC |
NC
NC |
NC
NC |
NC |
|
NC |
NC
NC |
NC |
NC |
|
|
|
| CCSD |
|
|
|
|
NC |
|
|
|
|
NC |
|
NC |
NC |
|
|
|
|
|
| CCSD=FULL |
|
|
|
|
NC |
|
|
|
|
NC |
|
NC |
NC |
|
|
|
|
|
| CCSD(T) |
|
|
|
|
NC |
NC |
|
NC |
|
|
|
NC |
NC |
|
|
|
|
|
| CCSD(T)=FULL |
|
|
|
|
NC |
|
|
|
|
|
|
|
NC |
|
|
|
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
cc-pV(T+d)Z |
daug-cc-pVTZ |
|---|