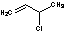
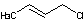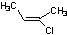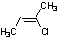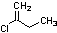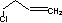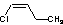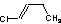Methods with standard basis sets
|
|
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
daug-cc-pVTZ |
| hartree fock |
HF |
0.0 a
-300.9 b
-320.7 c
-326.2 d
-307.6 e
-291.9 f
-302.9 g
-307.1 h |
0.0 a
-285.3 b
-276.6 c
-281.5 d
-270.2 e
-272.1 f
-258.1 g
-261.7 h |
0.0 a
-283.3 b
-284.5 c
-290.0 d
-279.0 e
-274.4 f
-271.8 g
-275.9 h |
0.0 a
-279.6 b
-276.1 c
-281.4 d
-266.9 e
-267.8 f
-257.3 g
-260.5 h |
0.0 a
-277.0 b
-282.1 c
-287.7 d
-274.9 e
-270.2 f
-268.9 g
-272.6 h |
0.0 a
-275.7 b
-280.5 c
-286.2 d
-273.5 e
-268.6 f
-267.2 g
-270.7 h |
0.0 a
-276.6 b
-277.2 c
-283.1 d
-271.7 e
-269.9 f
-264.4 g
-268.3 h |
0.0 a
-276.2 b
-276.9 c
-282.6 d
-270.9 e
-269.4 f
-263.9 g
-267.4 h |
0.0 a
-275.2 b
-275.2 c
-280.8 d
-269.3 e
-268.1 f
-262.0 g
-265.4 h |
0.0 a
-273.3 b
-279.7 c
-284.9 d
-272.7 e
-266.3 f
-266.0 g
-269.3 h |
NC
NC
NC
NC
NC
NC
NC |
0.0 a
-275.4 b
-274.6 c
-280.5 d
-269.4 e
-269.0 f
-261.9 g
-265.8 h |
0.0 a
-274.6 b
-277.4 c
-283.4 d
-270.9 e
-267.5 f
-264.1 g
-267.3 h |
0.0 a
-273.7 b
-276.2 c
-281.8 d
-270.6 e
-267.5 f
-263.4 g
-266.9 h |
0.0 a
-275.0 b
-273.1 c
-278.8 d
-268.7 e
-268.0 f
-260.4 g
-263.5 h |
0.0 a
-273.5 b
-275.4 c
-281.0 d
-269.8 e
-267.4 f
-262.6 g
-266.1 h |
0.0 a
-273.5 b
-275.4 c
-281.0 d
-269.8 e
-267.4 f
-262.6 g
-266.1 h |
| density functional |
LSDA |
0.0 a
-271.1 b
-302.1 c
-307.8 d
-285.7 e
-260.5 f
-278.9 g
-279.1 h |
NC
NC
NC
NC
NC
NC
NC |
0.0 a
-260.3 b
-279.5 c
-286.4 d
-270.1 e
-248.2 f
-258.9 g
-256.7 h |
0.0 a
-255.2 b
-271.0 c
-277.4 d
-258.3 e
-239.4 f
-244.6 g
-242.4 h |
0.0 a
-251.4 b
-276.9 c
-283.3 d
-264.7 e
-240.2 f
-254.7 g
-252.8 h |
0.0 a
-250.5 b
-276.1 c
-282.6 d
-264.2 e
-239.4 f
-254.0 g
-251.7 h |
0.0 a
-251.2 b
-272.9 c
-279.5 d
-262.4 e
-239.5 f
-251.1 g
-249.7 h |
0.0 a
-252.1 b
-272.0 c
-278.6 d
-262.1 e
-239.9 f
-250.9 g
-248.9 h |
0.0 a
-251.6 b
-271.6 c
-278.0 d
-261.5 e
-239.0 f
-249.7 g
-247.4 h |
0.0 a
-249.6 b
-277.1 c
-283.2 d
-265.3 e
-237.9 f
-253.9 g
-251.4 h |
|
0.0 a |
0.0 a
-249.3 b
-274.2 c
-280.9 d
-263.3 e
-238.5 f
-252.4 g
-250.0 h |
0.0 a
-249.5 b
-273.8 c
-280.3 d
-263.5 e
-238.6 f
-252.2 g
-249.8 h |
0.0 a
-250.0 b
-271.4 c
-278.2 d
-262.6 e
-238.8 f
-250.0 g
-247.3 h |
0.0 a |
|
| BLYP |
0.0 a
-264.7 b
-287.3 c
-292.1 d
-272.5 e
-252.8 f
-266.9 g
-271.0 h |
0.0 a
-255.5 b
-255.0 c
-259.9 d
-245.0 e
-239.0 f
-232.8 g
-236.2 h |
0.0 a
-254.3 b
-263.1 c
-268.2 d
-253.4 e
-242.5 f
-246.1 g
-249.7 h |
0.0 a
-249.7 b
-253.8 c
-258.7 d
-241.6 e
-233.7 f
-231.7 g
-234.7 h |
0.0 a
-247.1 b
-261.5 c
-266.6 d
-249.5 e
-236.2 f
-242.9 g
-246.2 h |
0.0 a
-246.6 b
-260.5 c
-265.8 d
-249.0 e
-238.9 f
-242.4 g
-245.5 h |
0.0 a
-247.0 b
-257.0 c
-262.5 d
-247.0 e
-236.0 f
-239.5 g
-243.2 h |
0.0 a
-246.8 b
-257.0 c
-262.2 d
-246.5 e
-238.7 f
-239.6 g
-242.9 h |
0.0 a
-246.7 b
-256.2 c
-261.3 d
-246.0 e
-235.1 f
-238.7 g
-242.0 h |
0.0 a
-245.9 b
-261.8 c
-266.7 d
-250.4 e
-234.7 f
-242.5 g
-245.5 h |
|
0.0 a |
0.0 a
-245.5 b
-258.6 c
-264.2 d
-248.6 e
-235.2 f
-241.6 g
-244.4 h |
0.0 a |
b
c
d
e
f
g
h |
0.0 a |
|
| B1B95 |
0.0 a
-263.1 b
-288.8 c
-293.6 d
-273.8 e
-252.8 f
-268.3 g
-270.9 h |
|
0.0 a
-268.7 b
-279.8 c
-285.9 d
-272.7 e
-258.4 f
-263.8 g
-264.7 h |
0.0 a
-266.1 b
-274.7 c
-280.6 d
-263.7 e
-252.6 f
-253.1 g
-253.6 h |
0.0 a
-258.8 b
-276.8 c
-282.8 d
-266.4 e
-250.1 f
-259.5 g
-260.3 h |
0.0 a
-285.6 b
-303.6 c
-309.8 d
-293.4 e
-276.6 f
-286.3 g
-286.9 h |
0.0 a
-260.3 b
-274.9 c
-281.2 d
-265.8 e
-251.2 f
-258.1 g
-259.0 h |
0.0 a
-261.3 b
-275.3 c
-281.4 d
-266.2 e
-251.9 f
-258.7 g
-259.2 h |
0.0 a
-260.8 b
-274.3 c
-280.5 d
-265.5 e
-251.0 f
-257.2 g
-257.8 h |
0.0 a
-256.6 b
-276.8 c
-282.4 d
-266.5 e
-247.7 f
-258.3 g
-259.0 h |
|
0.0 a |
0.0 a
-257.2 b
-274.4 c
-281.0 d
-265.3 e
-248.6 f
-257.0 g
-257.7 h |
0.0 a
-256.5 b
-273.3 c
-279.4 d
-264.7 e
-248.2 f
-256.2 g
-257.0 h |
0.0 a
-270.9 c
-277.4 d
-263.7 e
-248.6 f
-254.3 g
-254.5 h |
0.0 a |
|
| B3LYP |
0.0 a
-274.4 b
-297.2 c
-302.2 d
-282.7 e
-263.7 f
-277.1 g
-280.7 h |
0.0 a
-265.3 b
-264.4 c
-269.5 d
-255.2 e
-249.9 f
-242.7 g
-245.4 h |
0.0 a
-263.6 b
-272.3 c
-277.6 d
-263.6 e
-252.8 f
-255.9 g
-259.0 h |
0.0 a
-258.9 b
-263.4 c
-268.5 d
-251.8 e
-244.5 f
-241.7 g
-244.1 h |
0.0 a
-256.0 b
-270.2 c
-275.6 d
-259.2 e
-246.5 f
-252.5 g
-255.3 h |
0.0 a
-255.3 b
-269.2 c
-274.7 d
-258.6 e
-245.8 f
-251.8 g
-254.3 h |
0.0 a
-255.9 b
-265.9 c
-271.6 d
-256.8 e
-246.3 f
-249.0 g
-252.0 h |
0.0 a
-255.7 b
-265.7 c
-271.2 d
-256.2 e
-245.9 f
-248.9 g
-251.5 h |
0.0 a
-255.4 b
-264.8 c
-270.2 d
-255.5 e
-245.2 f
-247.8 g
-250.3 h |
0.0 a
-254.2 b
-270.0 c
-275.1 d
-259.5 e
-244.3 f
-251.5 g
-253.8 h |
NC
NC
NC
NC
NC
NC
NC |
0.0 a
-255.7 b
-265.1 c
-271.0 d
-256.4 e
-248.3 g
-251.1 h |
0.0 a
-254.1 b
-267.0 c
-272.8 d
-257.6 e
-245.0 f
-250.3 g
-252.5 h |
0.0 a
-254.5 b
-266.9 c
-272.5 d
-257.8 e
-245.4 f
-250.1 g
-252.7 h |
0.0 a
-254.7 b
-264.1 c
-269.8 d
-256.7 e
-245.3 f
-247.9 g
-249.9 h |
0.0 a
-254.2 b
-265.7 c
-271.2 d
-256.7 e
-245.1 f
-248.9 g
-251.5 h |
|
| B3LYPultrafine |
|
0.0 a |
|
|
0.0 a
-255.9 b
-270.1 c
-275.5 d
-259.1 e
-246.4 f
-252.4 g
-255.1 h |
0.0 a |
0.0 a |
0.0 a |
|
|
|
0.0 a |
0.0 a |
0.0 a
-254.3 b |
0.0 a |
0.0 a
-254.0 b
-265.5 c
-271.1 d
-256.5 e
-245.0 f
-248.8 g
-251.3 h |
|
| B3PW91 |
0.0 a
-275.0 b
-299.4 c
-304.5 d
-284.3 e
-264.5 f
-279.1 g
-282.8 h |
0.0 a
-266.9 b
-267.5 c
-273.2 d
-258.7 e
-251.3 f
-245.6 g
-247.8 h |
0.0 a
-264.8 b
-275.2 c
-281.0 d
-266.9 e
-254.0 f
-258.8 g
-261.4 h |
0.0 a
-259.9 b
-267.4 c
-272.8 d
-255.5 e
-246.0 f
-245.3 g |
0.0 a
-256.3 b
-273.3 c
-279.0 d
-262.1 e
-247.3 f
-255.5 g
-258.0 h |
0.0 a
-255.5 b
-272.5 c
-278.3 d
-261.5 e
-246.4 f
-254.7 g
-256.9 h |
0.0 a
-256.2 b
-269.9 c
-275.8 d
-260.3 e
-252.5 g
-255.1 h |
0.0 a
-256.1 b
-269.2 c
-275.0 d
-259.5 e
-246.7 f
-252.0 g
-254.1 h |
0.0 a
-255.6 b
-268.5 c
-274.2 d
-258.8 e
-245.9 f
-250.8 g
-252.8 h |
0.0 a
-253.8 b
-272.9 c
-278.2 d
-261.9 e
-244.6 f
-254.3 g
-256.3 h |
|
0.0 a |
0.0 a
-254.3 b
-270.5 c
-276.6 d
-260.2 e
-245.4 f
-252.9 g
-254.8 h |
0.0 a
-253.8 b
-270.2 c
-275.9 d
-260.3 e
-245.3 f
-252.7 g
-254.8 h |
0.0 a
5.7 b
-7.4 c
-13.4 d
0.9 e
14.7 f
9.7 g
8.1 h |
0.0 a |
|
| mPW1PW91 |
b
c
d
e
f
g
h |
0.0 a
-264.6 b
-265.0 c
-270.8 d
-256.8 e
-249.2 f
-243.1 g
-244.8 h |
0.0 a
-267.5 b
-277.7 c
-283.7 d
-270.0 e
-256.8 f
-261.4 g
-263.5 h |
0.0 a
-262.5 b
-269.9 c
-275.4 d
-258.5 e
-248.8 f
-247.8 g
-249.5 h |
0.0 a
-253.8 b
-270.6 c
-276.4 d
-259.9 e
-244.9 f
-252.8 g
-254.9 h |
0.0 a
-253.0 b
-269.7 c
-275.6 d
-259.3 e
-244.0 f
-252.0 g
-253.8 h |
0.0 a
-253.8 b
-267.1 c
-273.1 d
-258.0 e
-249.8 g
-252.0 h |
0.0 a
-253.6 b
-266.6 c
-272.4 d
-257.3 e
-244.4 f
-249.4 g
-251.2 h |
0.0 a
-258.0 b
-270.7 c
-276.4 d
-261.4 e
-248.3 f
-253.0 g
-254.7 h |
0.0 a
-256.2 b
-275.0 c
-280.5 d
-264.5 e
-247.1 f
-256.6 g
-258.2 h |
|
0.0 a |
0.0 a
-251.6 b
-267.7 c
-273.8 d
-257.7 e
-242.7 f
-249.9 g
-251.4 h |
0.0 a
-256.1 b
-272.2 c
-278.1 d
-262.8 e
-247.6 f
-254.8 g
-256.5 h |
0.0 a
-257.1 b
-269.8 c
-275.8 d
-261.8 e
-248.1 f
-252.6 g
-253.9 h |
0.0 a |
|
| M06-2X |
0.0 a |
0.0 a |
0.0 a
-273.2 b
-281.8 c
-277.5 e
-264.3 f
-264.1 g
-266.0 h |
0.0 a |
0.0 a
-264.3 b
-278.3 c
-284.7 d
-271.6 e
-260.8 g
-261.5 h |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
|
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
|
| PBEPBE |
0.0 a
-267.2 b
-293.3 c
-298.4 d
-277.8 e
-255.9 f
-272.0 g
-275.2 h |
0.0 a
-259.1 b
-261.8 c
-267.5 d
-252.4 e
-242.6 f
-238.6 g
-240.4 h |
0.0 a
-257.3 b
-269.6 c
-275.4 d
-260.6 e
-245.5 f
-251.9 g
-254.0 h |
0.0 a
-252.3 b
-261.4 c
-266.9 d
-249.1 e
-237.1 f
-238.2 g
-239.9 h |
0.0 a
-248.7 b
-268.0 c
-273.7 d
-255.9 e
-238.6 f
-248.7 g
-250.7 h |
0.0 a
-248.0 b
-267.2 c
-273.0 d
-255.5 e
-237.8 f
-248.1 g
-249.8 h |
0.0 a
-248.7 b
-264.0 c
-270.0 d
-253.9 e
-238.2 f
-245.5 g
-247.8 h |
0.0 a
-248.6 b
-263.6 c
-269.4 d
-253.2 e
-237.9 f
-245.2 g
-247.1 h |
0.0 a
-248.3 b
-263.0 c
-268.7 d
-252.6 e
-237.1 f
-244.2 g
-245.9 h |
0.0 a
-246.8 b
-268.0 c
-273.4 d
-256.4 e
-236.4 f
-248.0 g
-249.6 h |
|
0.0 a |
0.0 a
-246.9 b
-265.3 c
-271.4 d
-254.6 e
-236.9 f
-246.8 g
-248.3 h |
0.0 a
-246.7 b
-264.9 c
-270.8 d
-254.6 e
-236.9 f
-246.4 g
-248.1 h |
0.0 a
-247.3 b
-262.4 c
-268.5 d
-253.6 e
-237.1 f
-244.3 g
-245.6 h |
0.0 a |
|
| PBEPBEultrafine |
|
0.0 a |
|
|
0.0 a
-248.6 b
-267.9 c
-273.5 d
-255.8 e
-238.5 f
-248.6 g
-250.6 h |
0.0 a |
0.0 a |
0.0 a |
|
|
|
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
|
| PBE1PBE |
0.0 a |
|
0.0 a |
0.0 a |
0.0 a
-258.9 b
-276.3 c
-282.1 d
-265.7 e
-250.0 f
-258.3 g
-260.0 h |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
|
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
|
| HSEh1PBE |
|
0.0 a
-269.7 b
-270.7 c
-276.5 d
-262.3 e
-254.0 f
-248.3 g
-249.6 h |
0.0 a |
0.0 a |
0.0 a
-258.7 b
-276.2 c
-282.0 d
-265.4 e
-249.6 f
-258.0 g
-259.6 h |
0.0 a |
0.0 a
4.4 b
-9.3 c
-15.4 d
-0.2 e
13.8 f
8.3 g
6.5 h |
0.0 a |
0.0 a |
0.0 a |
|
0.0 a |
0.0 a |
0.0 a
-256.1 b
-272.6 c
-278.5 d
-263.3 e
-247.4 f
-254.9 g
-256.1 h |
0.0 a |
0.0 a |
|
| TPSSh |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a
-256.5 b
-271.7 c
-277.3 d
-260.9 e
-247.0 f
-253.9 g
-256.5 h |
0.0 a |
0.0 a
-256.4 b
-268.1 c
-274.0 d
-258.9 e
-246.9 f
-251.0 g
-253.7 h |
0.0 a |
0.0 a |
0.0 a
-253.8 b
-271.2 c
-276.6 d
-260.4 e
-244.3 f
-252.7 g
-254.8 h |
|
0.0 a |
0.0 a |
0.0 a
-253.7 b
-268.3 c
-274.0 d
-258.7 e
-244.8 f
-250.9 g
-253.1 h |
0.0 a |
0.0 a |
|
| wB97X-D |
0.0 a |
0.0 a |
0.0 a
-270.4 b
-278.2 c
-283.5 d
-272.9 e
-260.4 f
-264.5 g
-265.2 h |
0.0 a |
0.0 a
-262.3 b
-276.3 c
-267.8 e
-253.5 f
-260.6 g
-261.1 h |
0.0 a |
0.0 a
-262.1 b
-272.4 c
-265.5 e
-253.1 f
-257.3 g
-257.9 h |
0.0 a |
0.0 a
-261.6 b
-271.4 c
-264.3 e
-251.9 f
-255.8 g
-255.7 h |
0.0 a |
|
0.0 a
-261.6 b
-271.3 c
-264.7 e
-252.9 f
-256.4 g
-256.8 h |
0.0 a
-222.3 b
-232.6 c
-225.7 e
-213.2 f
-217.5 g
-218.1 h |
0.0 a
-259.7 b
-272.6 c
-265.5 e
-251.1 f
-257.3 g
-257.4 h |
0.0 a |
0.0 a
-259.5 b
-271.7 c
-264.7 e
-251.0 f
-256.6 g
-256.5 h |
|
| B97D3 |
|
0.0 a
-13.4 b
-11.4 c
-16.8 d
-3.8 e
3.8 f
11.0 g
9.3 h |
|
|
0.0 a
-2.7 b
-17.1 c
-22.8 d
-7.3 e
8.5 f
1.6 g
-0.0 h |
|
0.0 a
-4.4 b
-15.0 c
-20.9 d
-7.0 e
7.0 f
3.0 g
1.1 h |
|
0.0 a
-4.8 b
-15.0 c
-20.6 d
-6.6 e
7.1 f
3.4 g
2.0 h |
|
0.0 a
-252.2 b
-265.2 c
-271.0 d
-257.0 e
-240.9 f
-246.7 g
-248.1 h |
0.0 a
-4.2 b
-14.5 c
-20.6 d
-6.6 e
7.0 f
3.5 g
1.9 h |
|
0.0 a
-2.5 b
-16.0 c
-21.8 d
-7.6 e
8.2 f
2.1 g
0.7 h |
|
0.0 a
-2.8 b
-15.2 c
-21.1 d
-7.0 e
7.9 f
2.8 g
1.4 h |
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
daug-cc-pVTZ |
|---|
| Moller Plesset perturbation |
MP2 |
0.0 a
-283.5 b
-304.2 c
-309.0 d
-291.6 e
-273.7 f
-284.7 g
-289.2 h |
0.0 a
-274.5 b
-273.4 c
-278.8 d
-267.9 e
-258.0 f
-252.0 g
-255.0 h |
0.0 a
-275.1 b
-280.3 c
-286.6 d
-275.1 e
-261.5 f
-262.6 g
-266.1 h |
0.0 a
-268.1 b
-271.0 c
-276.8 d
-263.0 e
-252.1 f
-248.9 g
-251.8 h |
0.0 a
-270.4 b
-281.5 c
-287.9 d
-273.9 e
-257.4 f
-262.6 g
-265.7 h |
0.0 a
-271.6 b
-282.4 c
-289.0 d
-275.8 e
-258.6 f
-264.2 g
-267.1 h |
0.0 a
-272.5 e |
0.0 a
-270.2 b
-276.6 c
-282.8 d
-271.1 e
-256.9 f
-258.1 g
-261.0 h |
0.0 a
-269.2 b
-275.1 c
-281.1 d
-270.1 e
-256.0 f
-257.1 g
-259.8 h |
0.0 a
-2.4 b
-15.3 c
-21.3 d
-8.2 e
10.5 f
4.9 g
2.9 h |
|
0.0 a
-269.8 b
-273.2 c
-280.0 d
-268.8 e
-257.0 f
-255.2 g
-258.4 h |
0.0 a
-266.8 b
-277.0 c
-283.2 d
-271.2 e
-254.7 f
-259.3 g
-261.7 h |
0.0 a
-266.6 b
-277.2 c
-283.5 d
-271.7 e
-249.1 f
-258.4 g
-259.9 h |
0.0 a |
0.0 a |
|
| MP2=FULL |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a
-271.0 b
-282.4 c
-288.9 d
-274.8 e
-257.7 f
-263.2 g
-266.2 h |
0.0 a
-272.4 b
-283.6 c
-290.2 d
-276.9 e
-258.9 f
-264.9 g
-267.7 h |
0.0 a
-273.0 b
-279.2 c
-285.7 d
-273.6 e
-258.6 f
-260.0 g
-263.7 h |
0.0 a
-270.8 b
-277.6 c
-284.0 d
-272.0 e
-257.2 f
-258.7 g
-261.4 h |
0.0 a
-269.8 b
-276.1 c
-282.3 d
-271.1 e
-256.4 f
-257.8 g
-260.3 h |
0.0 a |
|
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
|
| MP3 |
|
|
|
|
0.0 a
-268.8 b
-278.4 c
-284.5 d
-271.7 e
-258.4 f
-262.7 g
-265.0 h |
|
0.0 a |
|
|
|
|
0.0 a |
0.0 a |
0.0 a |
|
|
|
| MP3=FULL |
|
0.0 a |
0.0 a |
0.0 a |
0.0 a
-269.3 b
-279.3 c
-285.4 d
-272.5 e
-258.6 f
-263.2 g
-265.4 h |
0.0 a |
0.0 a
-272.2 b
-277.0 c
-283.0 d
-272.0 e
-260.5 f
-261.2 g
-264.1 h |
0.0 a |
0.0 a |
0.0 a |
|
0.0 a |
0.0 a |
|
0.0 a |
|
|
| MP4 |
|
0.0 a |
|
|
0.0 a |
|
|
|
|
|
|
0.0 a |
0.0 a |
|
0.0 a |
|
|
| MP4=FULL |
|
0.0 a |
|
|
0.0 a |
|
|
|
|
|
|
|
0.0 a |
|
0.0 a |
|
|
| B2PLYP |
|
0.0 a |
0.0 a |
0.0 a |
0.0 a
-262.8 b
-275.3 c
-281.1 d
-266.0 e
-257.6 g
-260.4 h |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
|
0.0 a |
0.0 a |
0.0 a
-260.2 b
-271.3 c
-277.1 d
-263.7 e
-250.2 f
-254.0 g
-256.2 h |
0.0 a |
0.0 a |
|
| B2PLYP=FULL |
|
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
|
0.0 a |
0.0 a |
0.0 a |
0.0 a |
|
|
| Configuration interaction |
CID |
|
0.0 a |
0.0 a |
0.0 a |
0.0 a
-273.8 b
-282.8 c
-288.7 d
-275.6 e
-264.3 f
-267.2 g
-270.0 h |
|
|
0.0 a |
|
|
|
|
|
|
|
|
|
| CISD |
|
0.0 a |
0.0 a |
0.0 a |
0.0 a
-273.7 b
-282.6 c
-288.5 d
-275.4 e
-264.1 f
-267.0 g
-269.7 h |
|
|
0.0 a |
|
|
|
|
|
|
|
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
daug-cc-pVTZ |
|---|
| Quadratic configuration interaction |
QCISD |
|
0.0 a
-270.5 b
-266.9 c
-272.0 d
-262.0 e
-256.0 f
-247.7 g
-250.3 h |
0.0 a |
0.0 a |
0.0 a |
0.0 a
-269.7 b |
0.0 a
-270.3 b |
0.0 a |
0.0 a
-267.3 b |
0.0 a |
|
0.0 a |
0.0 a |
0.0 a |
0.0 a |
|
|
| QCISD(T) |
|
|
|
|
0.0 a |
|
|
|
|
|
|
0.0 a |
0.0 a |
|
0.0 a |
|
|
| QCISD(T)=FULL |
|
|
|
|
0.0 a |
|
|
|
|
|
|
|
0.0 a |
|
|
|
|
| Coupled Cluster |
CCD |
|
0.0 a |
0.0 a |
0.0 a |
0.0 a
-267.8 b
-276.8 c
-270.3 e
-257.6 f
-261.1 g
-263.6 h |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
0.0 a |
|
0.0 a |
0.0 a |
0.0 a |
0.0 a |
|
|
| CCSD |
|
|
|
|
0.0 a |
|
|
|
|
0.0 a |
|
0.0 a |
0.0 a |
|
0.0 a |
|
|
| CCSD=FULL |
|
|
|
|
0.0 a |
|
|
|
|
|
|
0.0 a |
0.0 a |
|
0.0 a |
|
|
| CCSD(T) |
|
|
|
|
0.0 a |
|
|
0.0 a |
|
|
|
0.0 a |
0.0 a |
|
0.0 a |
|
|
| CCSD(T)=FULL |
|
|
|
|
0.0 a |
|
|
|
|
|
|
|
0.0 a |
|
|
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
daug-cc-pVTZ |
|---|