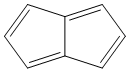
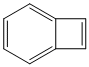
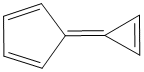
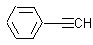Methods with standard basis sets
|
|
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
cc-pV(T+d)Z |
daug-cc-pVTZ |
| hartree fock |
HF |
49.5 a
89.0 b
265.2 c
0.0 d |
125.3 a
160.3 b
304.3 c
0.0 d |
125.3 a
160.3 b
304.3 c
0.0 d |
115.8 a
146.9 b
287.7 c
0.0 d |
86.3 a
112.8 b
223.0 c
0.0 d |
86.0 a
112.5 b
221.3 c
0.0 d |
87.5 a
115.6 b
220.0 c
0.0 d |
89.3 a
118.5 b
225.2 c
0.0 d |
90.4 a
119.7 b
223.2 c
0.0 d |
86.6 a
113.8 b
216.8 c
0.0 d |
0.0 d |
96.2 a
125.6 b
226.7 c
0.0 d |
82.4 a
111.1 b
218.6 c
0.0 d |
90.4 a
119.2 b
218.7 c
0.0 d |
215.2 c
0.0 d |
216.7 c
0.0 d |
0.0 d |
NC |
| density functional |
LSDA |
11.0 b
202.8 c
0.0 d |
44.9 a
82.3 b
234.9 c
0.0 d |
82.3 b
234.9 c
0.0 d |
69.6 b
213.5 c
0.0 d |
11.6 a
42.2 b
168.9 c
0.0 d |
11.9 a
42.4 b
167.7 c
0.0 d |
15.5 a
47.0 b
167.6 c
0.0 d |
26.4 a
57.2 b
178.6 c
0.0 d |
27.2 a
58.0 b
176.1 c
0.0 d |
15.6 a
47.0 b
167.7 c
0.0 d |
|
NC
NC |
10.5 a
43.0 b
168.5 c
0.0 d |
26.4 a
57.8 b
173.9 c
0.0 d |
168.8 c
0.0 d |
NC |
0.0 d |
|
| BLYP |
22.6 a
51.8 b
225.5 c
0.0 d |
89.4 a
120.2 b
258.6 c
0.0 d |
89.4 a
120.2 b
258.6 c
0.0 d |
84.2 a
112.8 b
241.7 c
0.0 d |
64.8 a
88.7 b
202.5 c
0.0 d |
65.0 a
89.0 b
201.4 c
0.0 d |
69.4 a
94.4 b
201.6 c
0.0 d |
79.2 a
103.5 b
211.6 c
0.0 d |
NC
NC
NC |
NC
NC
NC |
|
NC
NC
NC |
65.6 a
91.3 b
203.7 c
0.0 d |
81.9 a
106.2 b
208.8 c
0.0 d |
|
|
0.0 d |
|
| B1B95 |
11.5 a
41.7 b
216.6 c
0.0 d |
83.1 a
115.6 b
256.1 c
0.0 d |
83.1 a
115.6 b
256.1 c
0.0 d |
77.0 a
106.6 b
238.7 c
0.0 d |
43.2 a
66.5 b
176.3 c
0.0 d |
77.2 a
100.5 b
210.3 c
0.0 d |
55.8 a
80.4 b
184.6 c
0.0 d |
62.1 a
87.4 b
193.4 c
0.0 d |
62.7 a
88.1 b
191.0 c
0.0 d |
53.8 a
78.3 b
182.2 c
0.0 d |
|
NC
NC
NC |
49.6 a
75.1 b
183.5 c
0.0 d |
51.3 a
77.6 b
178.0 c
0.0 d |
173.9 c
0.0 d |
176.7 c
0.0 d |
0.0 d |
|
| B3LYP |
23.5 a
54.0 b
232.5 c
0.0 d |
92.5 a
123.8 b
266.3 c
0.0 d |
92.5 a
123.8 b
266.3 c
0.0 d |
85.4 a
114.1 b
248.7 c
0.0 d |
64.3 a
87.9 b
203.9 c
0.0 d |
64.4 a
88.0 b
202.6 c
0.0 d |
67.9 a
92.7 b
202.5 c
0.0 d |
76.7 a
101.3 b
212.0 c
0.0 d |
77.5 a
102.2 b
209.8 c
0.0 d |
70.6 a
94.3 b
203.5 c
0.0 d |
|
81.5 a
106.4 b
211.4 c
0.0 d |
64.1 a
89.5 b
204.1 c
0.0 d |
79.0 a
103.5 b
208.4 c
0.0 d |
203.5 c
0.0 d |
78.4 a
103.4 b
206.8 c
0.0 d |
0.0 d |
|
| B3LYPultrafine |
|
|
|
|
NC
NC
NC |
|
|
|
|
|
|
|
NC
NC
NC |
NC
NC
NC |
NC |
206.9 c
0.0 d |
|
|
| B3PW91 |
10.8 a
42.1 b
221.1 c
0.0 d |
74.7 a
107.8 b
251.7 c
0.0 d |
74.7 a
107.8 b
251.7 c
0.0 d |
68.3 a
98.2 b
233.4 c
0.0 d |
46.8 a
71.0 b
186.2 c
0.0 d |
46.9 a
71.1 b
184.8 c
0.0 d |
49.3 a
74.7 b
184.4 c
0.0 d |
56.0 a
81.6 b
192.6 c
0.0 d |
NC
NC
NC |
NC
NC
NC |
|
NC
NC
NC |
44.9 a
71.2 b
184.7 c
0.0 d |
57.3 a
83.1 b
188.2 c
0.0 d |
|
|
0.0 d |
|
| mPW1PW91 |
8.7 a
40.5 b
222.1 c
0.0 d |
72.4 a
106.0 b
252.4 c
0.0 d |
72.4 a
106.0 b
252.4 c
0.0 d |
66.0 a
96.4 b
234.1 c
0.0 d |
44.2 a
68.6 b
185.3 c
0.0 d |
44.3 a
68.7 b
183.8 c
0.0 d |
46.6 a
72.4 b
183.4 c
0.0 d |
52.7 a
78.7 b
191.2 c
0.0 d |
NC
NC
NC |
NC
NC
NC |
|
NC
NC
NC |
42.0 a
68.5 b
183.5 c
0.0 d |
54.0 a
80.2 b
186.9 c
0.0 d |
NC |
NC |
0.0 d |
|
| M06-2X |
NC
NC
NC |
NC
NC
NC |
88.8 a
119.9 b
262.2 c
0.0 d |
NC
NC
NC |
64.6 a
85.6 b
191.3 c
0.0 d |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
|
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC |
NC |
|
|
| PBEPBE |
1.7 a
32.4 b
210.1 c
0.0 d |
62.9 a
96.5 b
238.9 c
0.0 d |
62.9 a
96.4 b
238.9 c
0.0 d |
58.8 a
89.4 b
221.4 c
0.0 d |
38.4 a
63.4 b
178.3 c
0.0 d |
38.6 a
63.6 b
177.0 c
0.0 d |
42.0 a
68.2 b
176.9 c
0.0 d |
48.9 a
74.9 b
185.3 c
0.0 d |
49.6 a
75.7 b
182.9 c
0.0 d |
42.1 a
67.7 b
177.2 c
0.0 d |
|
NC
NC
NC |
37.4 a
64.3 b
177.6 c
0.0 d |
50.2 a
76.8 b
181.7 c
0.0 d |
NC |
NC |
0.0 d |
|
| PBEPBEultrafine |
|
|
|
|
38.3 a
63.4 b
178.2 c
0.0 d |
|
|
|
|
|
|
|
NC
NC
NC |
NC
NC
NC |
NC |
NC |
|
|
| PBE1PBE |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
40.9 a
65.2 b
181.8 c
0.0 d |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
|
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC |
NC |
|
|
| HSEh1PBE |
NC
NC
NC |
72.7 a
106.2 b
251.7 c
0.0 d |
NC
NC
NC |
NC
NC
NC |
43.9 a
68.0 b
183.3 c
0.0 d |
NC
NC
NC |
0.0 d |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
|
NC
NC
NC |
NC
NC
NC |
54.2 a
79.9 b
185.1 c
0.0 d |
NC |
NC |
|
|
| TPSSh |
|
|
|
|
45.4 a
68.1 b
0.0 d |
|
48.0 a
71.9 b
0.0 d |
|
|
49.0 a
72.3 b
179.6 c
0.0 d |
|
|
|
55.4 a
80.1 b
0.0 d |
|
|
|
|
| wB97X-D |
|
|
83.8 a
113.9 b
263.5 c
0.0 d |
|
53.7 a
73.9 b
197.6 c
0.0 d |
|
56.4 a
78.0 b
196.0 c
0.0 d |
|
63.8 a
85.7 b
201.3 c
0.0 d |
|
|
67.7 a
90.1 b
203.1 c
0.0 d |
56.4 a
78.0 b
196.0 c
0.0 d |
63.4 a
86.0 b
197.8 c
0.0 d |
|
63.3 a
86.2 b
196.8 c
0.0 d |
|
|
| B97D3 |
|
84.6 a
118.2 b
257.6 c
0.0 d |
|
|
60.7 a
86.4 b
194.5 c
0.0 d |
|
63.8 a
90.6 b
192.8 c
0.0 d |
|
71.2 a
98.1 b
199.4 c
0.0 d |
|
73.3 a
100.1 b
195.7 c
0.0 d |
76.3 a
103.3 b
202.3 c
0.0 d |
|
74.1 a
100.3 b
198.5 c
0.0 d |
|
73.5 a
100.3 b
197.1 c
0.0 d |
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
cc-pV(T+d)Z |
daug-cc-pVTZ |
|---|
| Moller Plesset perturbation |
MP2 |
83.1 a
127.4 b
308.9 c
0.0 d |
128.2 a
169.8 b
320.5 c
0.0 d |
128.2 a
169.8 b
320.5 c
0.0 d |
119.7 a
301.4 c
0.0 d |
71.3 a
101.1 b
225.1 c
0.0 d |
69.4 a
99.1 b
222.9 c
0.0 d |
0.0 d |
72.4 a
102.8 b
225.8 c
0.0 d |
71.8 a
101.9 b
222.8 c
0.0 d |
65.5 a
97.0 b
218.2 c
0.0 d |
|
|
63.7 a
95.5 b
222.1 c
0.0 d |
70.1 a
100.8 b
220.7 c
0.0 d |
c |
217.8 c
0.0 d |
0.0 d |
|
| MP2=FULL |
73.6 a
117.9 b
299.3 c
0.0 d |
127.9 a
169.7 b
320.3 c
0.0 d |
127.9 a
169.7 b
320.3 c
0.0 d |
119.5 a
301.3 c
0.0 d |
71.2 a
101.1 b
224.9 c
0.0 d |
69.5 a
99.3 b
222.8 c
0.0 d |
0.0 d |
71.9 a
102.4 b
225.2 c
0.0 d |
NC
NC
NC |
NC
NC
NC |
|
|
63.6 a
95.6 b
221.9 c
0.0 d |
71.8 a
101.8 b
219.8 c
0.0 d |
NC |
NC |
|
|
| MP3 |
|
|
|
|
NC
NC
NC |
|
|
|
|
|
|
|
|
|
|
|
|
|
| MP3=FULL |
|
|
|
|
54.4 a
87.7 b
212.3 c
0.0 d |
|
55.0 a
0.0 d |
|
|
|
|
|
|
|
|
|
|
|
| MP4 |
|
NC |
|
|
NC
NC |
|
|
|
NC |
|
|
|
NC |
|
|
|
|
|
| MP4=FULL |
|
NC |
|
|
NC
NC |
|
|
|
|
|
|
|
NC |
|
|
|
|
|
| B2PLYP |
NC |
NC |
NC |
NC |
214.5 c
0.0 d |
NC |
NC |
NC |
NC |
NC |
|
|
NC |
215.2 c
0.0 d |
NC |
NC |
|
|
| B2PLYP=FULL |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC |
|
|
NC |
NC |
NC |
NC |
|
|
| Configuration interaction |
CID |
|
114.9 a
154.6 b
303.5 c
0.0 d |
NC
NC
NC |
NC
NC
NC |
69.3 a
97.8 b
215.1 c
0.0 d |
|
|
NC
NC
NC |
|
|
|
|
|
|
|
|
|
|
| CISD |
|
114.6 a
154.7 b
303.3 c
0.0 d |
NC
NC
NC |
NC
NC
NC |
69.2 a
98.0 b
215.4 c
0.0 d |
|
|
NC
NC
NC |
|
|
|
|
|
|
|
|
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
cc-pV(T+d)Z |
daug-cc-pVTZ |
|---|
| Quadratic configuration interaction |
QCISD |
0.0 d |
101.8 a
148.4 b
303.8 c
0.0 d |
101.8 a
148.4 b
303.8 c
0.0 d |
94.4 a
136.4 b
286.5 c
0.0 d |
58.6 a
92.7 b
218.7 c
0.0 d |
56.9 a
90.8 b
216.7 c
0.0 d |
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC |
|
|
51.5 a
87.1 b
214.6 c
0.0 d |
|
|
|
|
|
| QCISD(T) |
|
|
|
|
NC
NC
NC |
|
|
|
|
|
|
|
|
|
|
|
|
|
| Coupled Cluster |
CCD |
|
103.7 a
148.5 b
306.9 c
0.0 d |
NC
NC
NC |
a
b
c |
59.6 a
92.3 b
219.3 c
0.0 d |
57.6 a
90.2 b
217.2 c
0.0 d |
|
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
|
|
NC
NC
NC |
NC |
|
|
|
|
| CCSD |
|
|
|
|
NC
NC
NC |
|
|
|
|
|
|
|
NC
NC
NC |
|
|
|
|
|
| CCSD=FULL |
|
|
|
|
NC
NC
NC |
|
|
|
|
|
|
|
NC
NC
NC |
|
|
|
|
|
| CCSD(T) |
|
|
|
|
NC
NC
NC |
|
|
|
|
|
|
|
NC |
|
|
|
|
|
| CCSD(T)=FULL |
|
|
|
|
NC |
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
cc-pV(T+d)Z |
daug-cc-pVTZ |
|---|