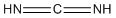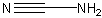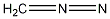Methods with predefined basis sets
| semi-empirical |
AM1 |
0.0 c |
| PM3 |
-56.4 a
128.6 b
0.0 c
0.4 d |
| PM6 |
91.1 b
0.0 c
-68.4 d |
| composite |
G1 |
-80.4 a
87.8 b
0.0 c
37.2 d |
| G2MP2 |
-84.0 a
82.2 b
0.0 c
34.5 d |
| G2 |
-83.0 a
65.0 b
0.0 c
35.6 d |
| G3 |
-82.5 a
74.0 b
0.0 c
37.1 d |
| G3B3 |
-77.9 a
75.3 b
0.0 c
42.2 d |
| G3MP2 |
-83.5 a
89.6 b
0.0 c
35.5 d |
| G4 |
-77.4 a
68.0 b
0.0 c
38.2 d |
| CBS-Q |
11.4 a
167.3 b
0.0 c
38.4 d |
Methods with standard basis sets
|
|
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
cc-pVQZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
aug-cc-pVQZ |
cc-pV(T+d)Z |
cc-pCVDZ |
cc-pCVTZ |
daug-cc-pVDZ |
daug-cc-pVTZ |
| hartree fock |
HF |
a
b
d |
70.4 a
353.5 b
0.0 c
226.6 d |
70.4 a
353.5 b
0.0 c
226.6 d |
-30.7 a
232.4 b
0.0 c
104.1 d |
|
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
20.6 a
0.0 c
170.2 d |
a
b
d |
a
b
d |
a
b
d |
21.3 a
0.0 c |
a
b
d |
a
b |
NC |
NC |
NC |
NC |
NC |
a
b
d |
| density functional |
LSDA |
a |
7.1 a
0.0 c |
7.1 a
0.0 c |
-83.4 a
0.0 c |
a
b |
a |
a |
a |
a |
a |
|
|
a |
a |
|
a |
|
|
|
|
|
|
|
| BLYP |
a
b
d |
16.7 a
244.8 b
0.0 c
127.2 d |
16.7 a
244.8 b
0.0 c
127.2 d |
-72.4 a
142.2 b
0.0 c
17.9 d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
NC |
a |
a
b |
a
b
d |
|
NC |
NC |
|
NC |
|
|
NC |
NC |
| B1B95 |
a
b
d |
22.0 a
261.1 b
0.0 c
147.8 d |
22.0 a
261.0 b
0.0 c
147.8 d |
18.1 a
238.2 b
0.0 c
121.8 d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
NC
NC |
a |
a
b
d |
a
b
d |
|
a
b
d |
a
b
d |
|
NC |
|
|
NC |
NC |
| B3LYP |
a
b |
29.1 a
272.8 b
0.0 c |
29.1 a
272.8 b
0.0 c
151.9 d |
-63.7 a
163.0 b
0.0 c
38.9 d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
NC |
a
b
d |
a
b
d |
a
b
d |
NC |
a
b
d |
a
b
d |
NC |
NC |
|
|
NC |
NC |
| B3LYPultrafine |
|
29.1 a
0.0 c |
|
|
a
b
d |
a |
a
d |
a |
|
NC |
NC |
a |
a |
a
d |
|
a |
a
b
d |
|
|
|
|
NC |
NC |
| B3PW91 |
a
b
d |
27.6 a
271.5 b
0.0 c
155.6 d |
27.6 a
271.5 b
0.0 c
155.6 d |
-64.8 a
159.2 b
0.0 c
40.0 d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
NC |
a |
a
b
d |
a
b
d |
|
NC |
NC |
|
NC |
|
|
NC |
NC |
| mPW1PW91 |
a
b
d |
30.4 a
275.8 b
0.0 c
161.4 d |
30.4 a
275.8 b
0.0 c
161.4 d |
-62.7 a
161.9 b
0.0 c
44.5 d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
NC |
a |
a
b
d |
a
b
d |
|
a |
a |
|
NC |
|
|
NC |
NC |
| M06-2X |
a |
46.2 a
0.0 c |
-44.2 a
195.4 b
0.0 c
89.6 d |
43.5 a
0.0 c |
a
b
d |
a |
a |
a |
a |
a |
a |
a |
a |
a |
|
a |
a |
|
|
|
|
NC |
NC |
| PBEPBE |
a
b
d |
14.8 a
241.9 b
0.0 c
130.5 d |
14.8 a
241.9 b
0.0 c
130.5 d |
-74.5 a
137.1 b
0.0 c
18.4 d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
NC |
a |
a
b
d |
a
b
d |
|
a |
a |
|
NC |
|
|
NC |
NC |
| PBEPBEultrafine |
|
14.8 a
0.0 c |
|
|
a
b
d |
a |
a |
a |
|
NC |
NC |
a |
a |
a |
|
a |
a |
|
|
|
|
NC |
NC |
| PBE1PBE |
a |
30.1 a
0.0 c |
30.1 a
0.0 c |
28.0 a
0.0 c |
a
b
d |
a |
a |
a |
a |
a |
NC |
a |
a |
a |
|
a |
a |
|
|
|
|
NC |
NC |
| HSEh1PBE |
a |
-61.1 a
0.0 c |
29.3 a
0.0 c |
27.2 a
0.0 c |
a |
a |
a |
a |
a |
a |
NC |
a |
a |
a |
|
a |
a |
|
|
|
|
NC |
NC |
| TPSSh |
NC |
24.8 a
0.0 c |
24.8 a
0.0 c |
22.9 a
0.0 c |
a
b
d |
a |
a
b
d |
a |
NC |
a
b
d |
NC |
a |
a |
a
b
d |
NC |
a |
a |
NC |
|
|
|
NC |
NC |
| wB97X-D |
NC |
NC |
-53.4 a
188.7 b
0.0 c
78.4 d |
NC |
a
b
d |
NC |
a
b
d |
NC |
a
b
d |
NC |
NC |
a
b
d |
a
b
d |
a
b
d |
NC |
NC |
a
b
d |
NC |
|
|
|
NC |
NC |
| B97D3 |
NC |
-70.9 a
164.7 b
0.0 c
45.8 d |
NC |
NC |
a
b
d |
NC |
a
b
d |
NC |
a
b
d |
NC |
a
b
d |
a
b
d |
NC |
a
b
d |
NC |
NC |
a
b
d |
NC |
|
|
|
NC |
a |
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
cc-pVQZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
aug-cc-pVQZ |
cc-pV(T+d)Z |
cc-pCVDZ |
cc-pCVTZ |
daug-cc-pVDZ |
daug-cc-pVTZ |
|---|
| Moller Plesset perturbation |
MP2 |
a
b
d |
62.4 a
273.7 b
0.0 c
182.3 d |
62.4 a
273.7 b
0.0 c
182.3 d |
-27.2 a
177.4 b
0.0 c
79.7 d |
a
b |
a
b
d |
a
b |
a
b |
a
b
d |
a
b
d |
NC |
a
b |
a
b
d |
|
NC |
a
d |
a
b
d |
NC |
NC |
NC |
NC |
NC |
NC |
| MP2=FULL |
a
b
d |
62.4 a
273.8 b
0.0 c
182.0 d |
62.4 a
273.8 b
0.0 c
182.0 d |
-27.2 a
177.5 b
0.0 c
79.4 d |
a
b |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
NC |
a |
a
b
d |
a
b
d |
NC |
a |
a
b |
NC |
NC |
NC |
NC |
NC |
NC |
| MP3 |
|
|
|
|
a
b
d |
|
a |
|
|
|
NC |
a |
a |
a |
|
|
|
|
|
|
|
NC |
NC |
| MP3=FULL |
|
NC |
NC |
NC |
a
b
d |
NC |
a
b
d |
NC |
NC |
NC |
NC |
a |
a |
a |
|
NC |
NC |
|
|
|
|
NC |
|
| MP4 |
0.0 c
144.4 d |
44.6 a
261.3 b
0.0 c
164.4 d |
|
|
a
b
d |
|
|
|
a
b
d |
0.0 c
126.4 d |
NC |
a |
a |
a |
|
a |
a |
|
|
|
|
NC |
NC |
| MP4=FULL |
|
44.5 a
0.0 c |
|
|
a |
|
|
|
a |
|
NC |
|
a |
a |
|
a |
a |
|
|
|
|
NC |
NC |
| B2PLYP |
a |
40.3 a
0.0 c |
40.3 a
0.0 c |
38.1 a
0.0 c |
a
b
d |
a |
a |
a |
a |
a |
NC |
a |
a |
a
b
d |
|
a |
a |
|
|
|
|
NC |
NC |
| B2PLYP=FULL |
a |
40.3 a
0.0 c |
40.3 a
0.0 c |
38.1 a
0.0 c |
a |
a |
a |
a |
a |
a |
NC |
a |
a |
a |
|
a |
a |
|
|
|
|
NC |
NC |
| Configuration interaction |
CID |
|
68.1 a
296.3 b
0.0 c
193.2 d |
68.1 a
296.3 b
0.0 c |
64.4 a
278.8 b
0.0 c |
a
b
d |
|
|
a
b |
|
0.0 c
149.7 d |
NC |
|
NC |
NC |
|
|
|
|
|
|
|
NC |
NC |
| CISD |
|
64.0 a
295.8 b
0.0 c
187.8 d |
64.0 a
295.8 b
0.0 c |
61.0 a
279.2 b
0.0 c |
a
b
d |
|
|
a
b |
|
0.0 c
147.5 d |
NC |
|
NC |
NC |
|
|
|
|
|
|
|
NC |
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
cc-pVQZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
aug-cc-pVQZ |
cc-pV(T+d)Z |
cc-pCVDZ |
cc-pCVTZ |
daug-cc-pVDZ |
daug-cc-pVTZ |
|---|
| Quadratic configuration interaction |
QCISD |
0.0 c
152.2 d |
61.6 a
277.0 b
0.0 c
172.4 d |
61.6 a
277.0 b
0.0 c |
-28.6 a
177.4 b
0.0 c
66.4 d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
d |
a
d |
NC |
a |
a
b
d |
a
d |
|
a |
a |
|
NC |
|
|
NC |
NC |
| QCISD(T) |
|
|
|
|
a
b
d |
|
|
NC |
0.0 c
131.7 d |
|
NC |
a |
a |
a |
|
a |
a |
|
|
|
|
NC |
NC |
| QCISD(T)=FULL |
|
|
|
|
a |
|
a |
|
|
|
NC |
|
a |
a |
NC |
a |
a |
NC |
|
|
|
NC |
NC |
| Coupled Cluster |
CCD |
0.0 c
184.5 d |
70.8 a
278.1 b
0.0 c
186.3 d |
70.8 a
278.1 b
0.0 c |
-21.4 a
175.9 b
0.0 c
80.6 d |
a
b
d |
a
b
d |
a
b
d |
a
b
d |
a
d |
a
d |
NC |
a |
a
b
d |
a
d |
|
a |
a |
|
NC |
|
|
NC |
NC |
| CCSD |
|
|
|
|
a
d |
NC |
NC |
NC |
NC
NC |
NC |
NC |
a |
a |
a |
NC |
a |
a |
|
|
|
|
NC |
NC |
| CCSD=FULL |
|
|
|
|
a |
|
|
|
|
NC |
NC |
a |
a |
a |
|
a |
a |
|
|
|
|
NC |
|
| CCSD(T) |
|
|
|
|
a
b
d |
NC |
NC |
NC |
22.2 a
0.0 c
132.3 d |
NC |
NC |
a |
a
b
d |
a
b
d |
NC |
a
b
d |
a
b
d |
NC |
|
NC |
NC |
NC |
NC |
| CCSD(T)=FULL |
|
|
|
|
a
d |
|
|
|
|
|
NC |
a |
a
d |
a |
NC |
a |
a
b |
NC |
|
NC |
NC |
NC |
NC |
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
cc-pVQZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
aug-cc-pVQZ |
cc-pV(T+d)Z |
cc-pCVDZ |
cc-pCVTZ |
daug-cc-pVDZ |
daug-cc-pVTZ |
|---|
Methods with effective core potentials (select basis sets)
|
|
CEP-31G |
CEP-31G* |
CEP-121G |
CEP-121G* |
LANL2DZ |
SDD |
cc-pVTZ-PP |
aug-cc-pVTZ-PP |
Def2TZVPP |
| hartree fock |
HF |
67.0 a
298.6 b
0.0 c
202.8 d |
a
b
d |
69.1 a
301.2 b
0.0 c
204.2 d |
a
b
d |
64.1 a
320.3 b
0.0 c
200.7 d |
64.1 a
320.0 b
0.0 c
200.9 d |
|
|
a
b
d |
| density functional |
BLYP |
|
|
|
|
|
|
|
|
NC |
| B1B95 |
0.0 c
137.0 d |
-69623.1 d |
|
|
|
|
|
|
NC |
| B3LYP |
27.2 a
229.0 b
0.0 c
132.3 d |
a
b
d |
30.0 a
232.2 b
0.0 c
134.2 d |
a
b
d |
24.0 a
247.8 b
0.0 c
128.2 d |
24.0 a
247.0 b
0.0 c
128.1 d |
|
|
a
b
d |
| B3LYPultrafine |
|
|
|
|
|
|
|
|
NC |
| B3PW91 |
|
|
|
|
|
|
|
|
NC |
| mPW1PW91 |
|
|
|
|
|
|
|
|
NC |
| M06-2X |
|
|
|
|
|
|
|
|
NC |
| PBEPBE |
|
|
|
|
|
|
|
|
a
b
d |
| PBEPBEultrafine |
|
|
|
|
|
|
|
|
NC |
| PBE1PBE |
|
|
|
|
|
|
|
|
NC |
| HSEh1PBE |
|
|
|
|
|
|
|
|
NC |
| TPSSh |
|
|
|
|
|
|
|
|
NC |
| wB97X-D |
-69667.6 a |
-69819.1 a |
-69686.2 a |
-69848.9 a |
NC |
NC |
|
|
NC |
| B97D3 |
|
|
|
|
|
|
|
|
NC |
| Moller Plesset perturbation |
MP2 |
62.7 a
230.6 b
0.0 c
171.6 d |
a
b
d |
64.4 a
232.7 b
0.0 c
172.6 d |
a
b
d |
60.1 a
258.3 b
0.0 c
170.4 d |
60.3 a
257.0 b
0.0 c
170.2 d |
|
|
a
b
d |
| MP2=FULL |
|
|
|
|
|
|
|
|
NC |
| MP3 |
|
|
|
|
|
|
|
|
NC |
| MP3=FULL |
|
|
|
|
|
|
|
|
NC |
| MP4=FULL |
|
|
|
|
|
|
|
|
NC |
| B2PLYP |
|
|
|
|
|
|
|
|
NC |
| B2PLYP=FULL |
|
|
|
|
|
|
|
|
NC |
| Configuration interaction |
CID |
|
|
|
|
|
|
|
|
NC |
| CISD |
|
|
|
|
|
|
|
|
NC |
| Quadratic configuration interaction |
QCISD |
|
|
|
|
|
|
|
|
NC |
| QCISD(T)=FULL |
|
|
|
|
|
|
|
|
NC |
| Coupled Cluster |
CCD |
|
|
|
|
|
|
|
|
NC |
| CCSD |
|
|
|
|
|
|
|
|
NC |
| CCSD=FULL |
|
|
|
|
|
|
|
|
NC |
| CCSD(T) |
|
|
|
|
|
|
|
|
NC |
| CCSD(T)=FULL |
|
|
|
|
|
|
|
|
NC |
NC = not calculated