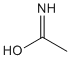
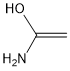
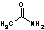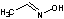Methods with standard basis sets
|
|
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
cc-pV(T+d)Z |
daug-cc-pVDZ |
daug-cc-pVTZ |
| hartree fock |
HF |
252.5 a
33.2 b
0.0 d
334.1 e |
254.3 a
74.3 b
0.0 d
437.3 e |
254.3 a
74.3 b
0.0 d
437.3 e |
267.6 a
75.7 b
0.0 d
453.8 e |
250.8 a
61.1 b
0.0 d
449.4 e |
256.8 a
55.2 b
0.0 d
449.3 e |
257.6 a
54.8 b
0.0 d |
252.2 a
65.7 b
0.0 d
453.8 e |
255.5 a
54.8 b
0.0 d
447.8 e |
254.4 a
54.0 b
0.0 d
449.4 e |
54.4 b
0.0 d
244.0 e |
255.8 a
55.2 b
0.0 d
447.8 e |
253.7 a
53.7 b
0.0 d
443.9 e |
255.9 a
53.2 b
0.0 d
449.0 e |
257.0 a
50.9 b
0.0 d
445.5 e |
255.5 a
52.8 b
0.0 d
450.1 e |
0.0 d
239.6 e |
NC |
255.5 a
52.7 b
0.0 d
450.1 e |
| density functional |
LSDA |
|
NC
NC |
|
|
NC
NC |
NC
NC |
NC |
NC
NC |
NC
NC |
NC
NC |
|
|
NC
NC |
NC
NC |
NC |
NC |
|
|
|
| BLYP |
210.8 a
29.4 b
0.0 d
285.1 e |
217.5 a
75.2 b
0.0 d |
217.5 a
75.2 b
0.0 d |
228.3 a
76.9 b
0.0 d |
217.0 a
56.0 b
0.0 d |
222.0 a
50.9 b
0.0 d |
227.7 a
52.7 b
0.0 d |
a
b |
225.2 a
53.6 b
0.0 d
388.3 e |
220.2 a
50.3 b
0.0 d |
NC |
NC |
220.0 a
51.1 b
0.0 d
379.8 e |
226.3 a
51.0 b
0.0 d |
49.1 b
0.0 d |
NC |
0.0 d
201.0 e |
NC |
NC |
| B1B95 |
229.8 a
35.7 b
0.0 d
312.5 e |
243.5 a
86.8 b
0.0 d
419.8 e |
243.5 a
86.8 b
0.0 d
419.8 e |
256.5 a
89.8 b
0.0 d
433.5 e |
231.9 a
56.6 b
0.0 d |
268.7 a
93.4 b
0.0 d |
250.3 a
60.9 b
0.0 d |
244.6 a
72.4 b
0.0 d
427.5 e |
248.0 a
61.8 b
0.0 d
421.9 e |
243.3 a
57.6 b
0.0 d
417.2 e |
NC |
NC
NC
NC |
242.8 a
57.8 b
0.0 d
412.9 e |
248.4 a
58.9 b
0.0 d
420.9 e |
240.8 a
48.1 b
0.0 d
408.4 e |
49.5 b
0.0 d
412.7 e |
0.0 d
226.9 e |
|
NC |
| B3LYP |
222.5 a
31.8 b
0.0 d
301.8 e |
229.8 a
77.2 b
0.0 d
402.4 e |
229.8 a
77.2 b
0.0 d
402.4 e |
240.8 a
78.2 b
0.0 d
413.7 e |
227.9 a
57.3 b
0.0 d |
233.0 a
51.9 b
0.0 d
403.2 e |
237.7 a
53.1 b
0.0 d |
231.9 a
63.9 b
0.0 d
412.4 e |
235.1 a
53.9 b
0.0 d
407.1 e |
231.2 a
51.2 b
0.0 d
404.2 e |
52.1 b
0.0 d
217.0 e |
NC
NC |
230.9 a
51.4 b
0.0 d
398.9 e |
a
b
e |
237.2 a
49.5 b
0.0 d |
237.2 a
50.9 b
0.0 d |
e |
NC |
NC |
| B3LYPultrafine |
|
NC
NC
NC |
|
|
227.9 a
57.3 b
0.0 d
403.2 e |
NC
NC
NC |
NC
NC |
NC
NC
NC |
|
NC |
NC |
NC
NC
NC |
NC
NC
NC |
236.2 a
51.5 b
0.0 d
406.8 e |
NC
NC |
51.0 b
0.0 d |
|
NC |
NC |
| B3PW91 |
226.6 a
34.0 b
0.0 d
309.4 e |
234.8 a
79.4 b
0.0 d
410.9 e |
234.8 a
79.4 b
0.0 d
410.9 e |
244.3 a
80.3 b
0.0 d
421.2 e |
231.2 a
58.6 b
0.0 d |
236.3 a
52.9 b
0.0 d
408.1 e |
240.2 a
53.7 b
0.0 d |
234.2 a
64.9 b
0.0 d
417.4 e |
237.6 a
54.4 b
0.0 d
411.9 e |
234.6 a
51.7 b
0.0 d
408.4 e |
NC |
NC
NC |
233.7 a
52.0 b
0.0 d
404.0 e |
238.3 a
51.4 b
0.0 d
410.1 e |
239.1 a
49.9 b
0.0 d |
51.1 b
0.0 d
410.6 e |
0.0 d
216.6 e |
NC |
NC |
| mPW1PW91 |
229.3 a
33.8 b
0.0 d
312.3 e |
232.8 a
75.0 b
0.0 d
410.3 e |
237.0 a
79.2 b
0.0 d
414.5 e |
247.0 a
80.2 b
0.0 d
425.2 e |
229.4 a
54.7 b
0.0 d |
234.8 a
49.0 b
0.0 d
408.1 e |
238.8 a
50.0 b
0.0 d |
236.2 a
64.9 b
0.0 d
421.1 e |
239.7 a
54.2 b
0.0 d
415.5 e |
236.9 a
51.6 b
0.0 d
412.1 e |
NC |
NC
NC
NC |
231.9 a
48.0 b
0.0 d
403.9 e |
236.7 a
47.6 b
0.0 d
410.0 e |
241.4 a
49.9 b
0.0 d |
NC
NC |
0.0 d
219.0 e |
NC |
NC |
| M06-2X |
NC
NC |
NC |
235.1 a
0.0 d |
NC
NC |
a
b |
NC
NC |
NC
NC |
NC
NC |
NC
NC |
a
b |
|
NC
NC |
NC
NC |
240.4 a
44.6 b
0.0 d |
NC |
a
b |
|
NC |
NC |
| PBEPBE |
215.5 a
31.4 b
0.0 d
296.1 e |
222.5 a
77.6 b
0.0 d |
222.5 a
77.6 b
0.0 d |
233.1 a
79.5 b
0.0 d |
221.1 a
57.3 b
0.0 d |
226.4 a
52.0 b
0.0 d |
231.6 a
53.5 b
0.0 d |
225.5 a
64.3 b
0.0 d |
228.8 a
54.0 b
0.0 d
395.8 e |
224.5 a
50.4 b
0.0 d
390.8 e |
b |
NC
NC |
223.8 a
51.4 b
0.0 d
387.2 e |
229.6 a
50.8 b
0.0 d |
230.7 a
49.5 b
0.0 d |
231.0 a
50.4 b
0.0 d |
0.0 d
206.6 e |
NC |
NC |
| PBEPBEultrafine |
|
NC
NC |
|
|
221.2 a
57.4 b
0.0 d |
NC
NC |
NC
NC |
NC
NC |
|
NC |
NC |
NC
NC |
NC
NC
NC |
NC
NC |
NC
NC |
NC
NC |
|
NC |
NC |
| PBE1PBE |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
233.3 a
58.5 b
0.0 d |
NC
NC |
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC |
NC
NC |
|
NC |
NC |
| HSEh1PBE |
NC
NC
NC |
236.3 a
79.5 b
0.0 d
414.6 e |
NC
NC
NC |
NC
NC
NC |
a
b
e |
NC
NC
NC |
242.2 a
53.6 b
0.0 d |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC |
NC
NC
NC |
NC
NC
NC |
a
b
e |
NC
NC |
NC
NC
NC |
|
NC |
NC |
| TPSSh |
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
222.6 a
57.6 b
0.0 d |
NC
NC
NC |
231.6 a
53.4 b
0.0 d |
NC
NC
NC |
NC |
51.0 b
0.0 d
203.3 e |
NC |
NC
NC
NC |
NC
NC
NC |
229.5 a
51.2 b
0.0 d
397.7 e |
NC
NC |
NC
NC
NC |
|
NC |
NC |
| wB97X-D |
NC |
NC
NC |
240.8 a
83.7 b
0.0 d |
NC
NC |
234.4 a
59.8 b
0.0 d |
NC
NC |
a
b |
NC
NC |
55.4 b
0.0 d |
NC |
NC |
a
b |
a
b
e |
241.2 a
0.0 d |
NC |
241.5 a
0.0 d |
|
NC |
|
| B97D3 |
NC |
77.0 b
0.0 d
212.7 e |
NC |
NC |
57.0 b
0.0 d
205.3 e |
NC |
b
e |
NC |
b
e |
NC |
a
b |
54.5 b
0.0 d
211.5 e |
NC |
b
e |
NC |
b
e |
|
NC |
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
cc-pV(T+d)Z |
daug-cc-pVDZ |
daug-cc-pVTZ |
|---|
| Moller Plesset perturbation |
MP2 |
232.0 a
39.2 b
0.0 d
315.5 e |
235.9 a
73.5 b
0.0 d |
235.9 a
73.5 b
0.0 d |
247.9 a
72.5 b
0.0 d |
237.6 a
55.4 b
0.0 d |
241.8 a
49.9 b
0.0 d |
a
b
e |
241.5 a
63.9 b
0.0 d |
47.8 b
0.0 d |
241.2 a
49.1 b
0.0 d |
NC |
239.9 a
48.7 b
0.0 d |
238.8 a
48.0 b
0.0 d |
242.8 a
0.0 d |
241.8 a
45.8 b
0.0 d |
b |
e |
NC |
NC |
| MP2=FULL |
NC
NC
NC |
235.9 a
73.6 b
0.0 d |
NC
NC |
NC
NC |
237.8 a
55.8 b
0.0 d |
242.3 a
50.5 b
0.0 d |
243.9 a
50.8 b
0.0 d |
242.2 a
64.1 b
0.0 d |
243.5 a
48.2 b
0.0 d |
244.2 a
51.2 b
0.0 d |
NC |
NC
NC |
239.0 a
48.3 b
0.0 d |
243.9 a
46.2 b
0.0 d
420.1 e |
NC
NC |
244.0 a
47.4 b
0.0 d |
|
NC |
NC |
| MP3 |
|
|
|
|
241.8 a
48.8 b
0.0 d |
|
207.4 a
3.9 b
0.0 d |
|
|
|
NC |
NC
NC |
NC
NC
NC |
NC
NC |
|
|
|
NC |
|
| MP3=FULL |
|
NC
NC
NC |
NC
NC
NC |
NC
NC |
242.0 a
49.3 b
0.0 d |
NC
NC |
247.8 a
44.7 b
0.0 d |
NC
NC |
NC
NC |
NC
NC |
NC |
NC
NC |
NC
NC
NC |
NC
NC |
NC
NC |
NC |
|
NC |
|
| MP4 |
|
NC
NC |
NC |
|
NC |
|
|
|
NC |
|
NC |
NC |
NC
NC |
|
NC |
NC |
|
NC |
|
| MP4=FULL |
|
NC
NC |
|
|
NC
NC |
|
|
|
NC
NC |
|
NC |
|
NC
NC |
NC
NC
NC |
NC
NC |
NC |
|
|
|
| B2PLYP |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC |
231.7 a
57.1 b
0.0 d |
NC
NC |
NC
NC |
NC
NC |
NC
NC
NC |
235.2 a
50.8 b
0.0 d |
NC |
NC
NC |
NC
NC
NC |
238.9 a
49.9 b
0.0 d |
NC
NC |
239.7 a
49.8 b
0.0 d |
|
NC |
NC |
| B2PLYP=FULL |
NC
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC |
NC
NC |
NC
NC |
NC
NC |
NC
NC |
NC
NC
NC |
NC
NC |
NC |
NC
NC |
NC
NC
NC |
NC
NC
NC |
NC
NC |
NC |
|
NC |
NC |
| Configuration interaction |
CID |
|
NC
NC
NC |
NC
NC
NC |
NC
NC |
245.2 a
55.4 b
0.0 d |
|
|
NC
NC |
|
|
NC |
|
NC |
NC |
|
|
|
NC |
|
| CISD |
|
NC
NC
NC |
NC
NC
NC |
NC
NC |
244.5 a
56.1 b
0.0 d |
|
|
NC
NC |
|
|
NC |
|
NC |
NC |
|
|
|
NC |
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
cc-pV(T+d)Z |
daug-cc-pVDZ |
daug-cc-pVTZ |
|---|
| Quadratic configuration interaction |
QCISD |
|
231.6 a
66.5 b
0.0 d |
NC
NC |
NC |
236.4 a
52.5 b
0.0 d |
241.1 a
47.8 b
0.0 d |
NC
NC |
239.9 a
60.7 b
0.0 d |
NC
NC |
241.2 a
47.9 b
0.0 d |
NC |
NC
NC |
NC
NC |
242.6 a
44.9 b
0.0 d |
NC
NC |
|
|
NC |
|
| QCISD(T) |
|
|
|
|
NC |
|
|
NC |
|
|
NC |
NC
NC |
NC
NC
NC |
NC |
NC
NC |
|
|
NC |
|
| QCISD(T)=FULL |
|
|
|
|
NC
NC |
|
NC
NC |
|
|
|
NC |
|
NC |
NC |
NC
NC |
|
|
NC |
|
| Coupled Cluster |
CCD |
|
NC
NC
NC |
NC
NC
NC |
NC
NC |
239.1 a
49.8 b
0.0 d |
NC
NC |
NC
NC |
NC
NC |
NC
NC |
NC
NC |
NC |
NC
NC |
NC
NC
NC |
NC
NC |
NC
NC |
|
|
NC |
|
| CCSD |
|
|
|
|
237.2 a
50.8 b
0.0 d |
NC |
NC |
NC |
NC |
241.9 a
46.5 b
0.0 d |
NC |
NC
NC |
NC
NC
NC |
243.3 a
43.6 b
0.0 d
408.3 e |
NC
NC |
|
|
|
|
| CCSD=FULL |
|
|
|
|
237.4 a
51.3 b
0.0 d |
|
|
|
|
244.7 a
48.8 b
0.0 d |
NC |
NC
NC |
NC
NC
NC |
244.5 a
44.3 b
0.0 d |
NC
NC |
|
|
NC |
|
| CCSD(T) |
|
|
|
|
NC |
NC |
NC |
NC |
NC |
NC |
NC |
NC
NC |
NC
NC
NC |
NC |
NC
NC |
|
|
NC |
|
| CCSD(T)=FULL |
|
|
|
|
NC |
|
|
|
|
|
|
NC |
NC
NC |
|
NC |
|
|
|
|
| |
STO-3G |
3-21G |
3-21G* |
6-31G |
6-31G* |
6-31G** |
6-31+G** |
6-311G* |
6-311G** |
6-31G(2df,p) |
6-311+G(3df,2p) |
TZVP |
cc-pVDZ |
cc-pVTZ |
aug-cc-pVDZ |
aug-cc-pVTZ |
cc-pV(T+d)Z |
daug-cc-pVDZ |
daug-cc-pVTZ |
|---|